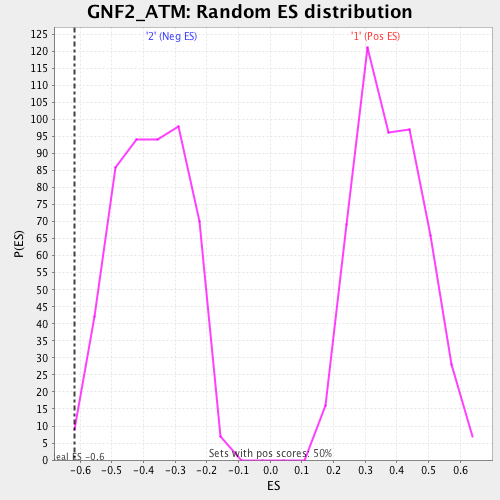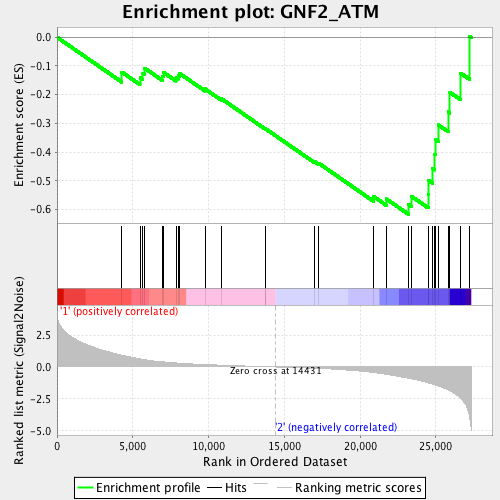
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | GNF2_ATM |
| Enrichment Score (ES) | -0.6175452 |
| Normalized Enrichment Score (NES) | -1.6383215 |
| Nominal p-value | 0.008 |
| FDR q-value | 0.10966606 |
| FWER p-Value | 0.495 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SYNRG | NA | 4275 | 0.913 | -0.1213 | No | ||
| 2 | GPR171 | NA | GPR171 Entrez, Source | G protein-coupled receptor 171 | 5470 | 0.624 | -0.1409 | No |
| 3 | OGT | NA | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 5663 | 0.585 | -0.1253 | No |
| 4 | ATHL1 | NA | ATHL1 Entrez, Source | ATH1, acid trehalase-like 1 (yeast) | 5782 | 0.563 | -0.1078 | No |
| 5 | RASGRP1 | NA | RASGRP1 Entrez, Source | RAS guanyl releasing protein 1 (calcium and DAG-regulated) | 6957 | 0.401 | -0.1353 | No |
| 6 | SFI1 | NA | SFI1 Entrez, Source | Sfi1 homolog, spindle assembly associated (yeast) | 7012 | 0.396 | -0.1219 | No |
| 7 | TRAF3IP3 | NA | TRAF3IP3 Entrez, Source | TRAF3 interacting protein 3 | 7845 | 0.313 | -0.1403 | No |
| 8 | RASGRP2 | NA | RASGRP2 Entrez, Source | RAS guanyl releasing protein 2 (calcium and DAG-regulated) | 7985 | 0.302 | -0.1337 | No |
| 9 | RAPGEF6 | NA | RAPGEF6 Entrez, Source | Rap guanine nucleotide exchange factor (GEF) 6 | 8083 | 0.294 | -0.1259 | No |
| 10 | PIK3IP1 | NA | 9760 | 0.190 | -0.1799 | No | ||
| 11 | LAT | NA | LAT Entrez, Source | linker for activation of T cells | 10843 | 0.140 | -0.2141 | No |
| 12 | ACAP1 | NA | 13721 | 0.025 | -0.3186 | No | ||
| 13 | GZMK | NA | GZMK Entrez, Source | granzyme K (granzyme 3; tryptase II) | 13731 | 0.025 | -0.3179 | No |
| 14 | DENND2D | NA | DENND2D Entrez, Source | DENN/MADD domain containing 2D | 16954 | -0.099 | -0.4321 | No |
| 15 | ITK | NA | ITK Entrez, Source | IL2-inducible T-cell kinase | 17238 | -0.112 | -0.4381 | No |
| 16 | HECA | NA | HECA Entrez, Source | headcase homolog (Drosophila) | 20877 | -0.435 | -0.5546 | No |
| 17 | PPWD1 | NA | PPWD1 Entrez, Source | peptidylprolyl isomerase domain and WD repeat containing 1 | 21726 | -0.576 | -0.5634 | No |
| 18 | ZNF266 | NA | ZNF266 Entrez, Source | zinc finger protein 266 | 23206 | -0.881 | -0.5835 | Yes |
| 19 | LDLRAP1 | NA | LDLRAP1 Entrez, Source | low density lipoprotein receptor adaptor protein 1 | 23391 | -0.924 | -0.5545 | Yes |
| 20 | BTN3A1 | NA | BTN3A1 Entrez, Source | butyrophilin, subfamily 3, member A1 | 24494 | -1.241 | -0.5468 | Yes |
| 21 | BTN3A2 | NA | BTN3A2 Entrez, Source | butyrophilin, subfamily 3, member A2 | 24522 | -1.253 | -0.4993 | Yes |
| 22 | CASP8 | NA | CASP8 Entrez, Source | caspase 8, apoptosis-related cysteine peptidase | 24765 | -1.335 | -0.4566 | Yes |
| 23 | ATM | NA | ATM Entrez, Source | ataxia telangiectasia mutated (includes complementation groups A, C and D) | 24929 | -1.388 | -0.4088 | Yes |
| 24 | DUSP2 | NA | DUSP2 Entrez, Source | dual specificity phosphatase 2 | 24949 | -1.396 | -0.3555 | Yes |
| 25 | RNF125 | NA | RNF125 Entrez, Source | ring finger protein 125 | 25145 | -1.482 | -0.3053 | Yes |
| 26 | TMC6 | NA | TMC6 Entrez, Source | transmembrane channel-like 6 | 25801 | -1.797 | -0.2598 | Yes |
| 27 | DEF6 | NA | DEF6 Entrez, Source | differentially expressed in FDCP 6 homolog (mouse) | 25919 | -1.866 | -0.1918 | Yes |
| 28 | CLEC2D | NA | CLEC2D Entrez, Source | C-type lectin domain family 2, member D | 26611 | -2.396 | -0.1244 | Yes |
| 29 | HMHA1 | NA | HMHA1 Entrez, Source | histocompatibility (minor) HA-1 | 27216 | -3.895 | 0.0041 | Yes |