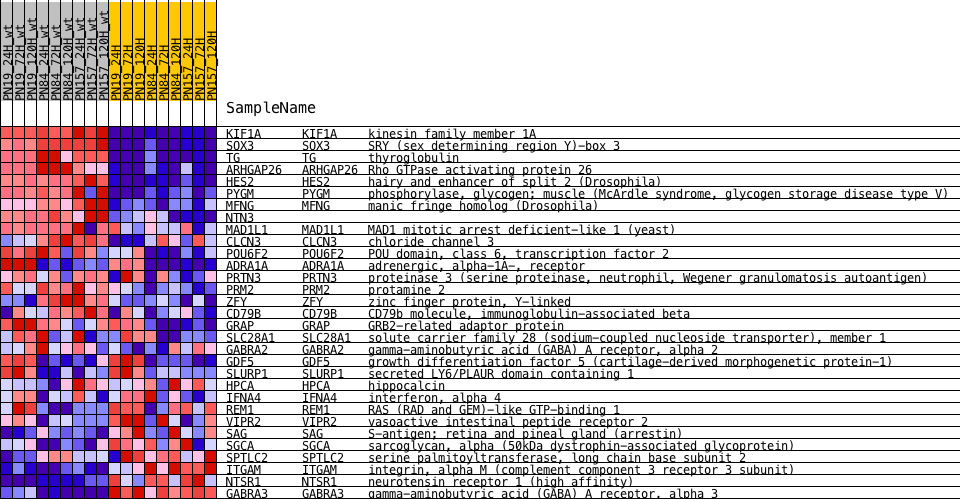
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | MODULE_489 |
| Enrichment Score (ES) | 0.5108886 |
| Normalized Enrichment Score (NES) | 1.7144445 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06923025 |
| FWER p-Value | 0.301 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KIF1A | NA | KIF1A Entrez, Source | kinesin family member 1A | 80 | 3.535 | 0.1479 | Yes |
| 2 | SOX3 | NA | SOX3 Entrez, Source | SRY (sex determining region Y)-box 3 | 241 | 3.192 | 0.2781 | Yes |
| 3 | TG | NA | TG Entrez, Source | thyroglobulin | 775 | 2.494 | 0.3650 | Yes |
| 4 | ARHGAP26 | NA | ARHGAP26 Entrez, Source | Rho GTPase activating protein 26 | 1765 | 1.843 | 0.4074 | Yes |
| 5 | HES2 | NA | HES2 Entrez, Source | hairy and enhancer of split 2 (Drosophila) | 1964 | 1.744 | 0.4745 | Yes |
| 6 | PYGM | NA | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 2652 | 1.443 | 0.5109 | Yes |
| 7 | MFNG | NA | MFNG Entrez, Source | manic fringe homolog (Drosophila) | 4627 | 0.832 | 0.4741 | No |
| 8 | NTN3 | NA | 6512 | 0.452 | 0.4243 | No | ||
| 9 | MAD1L1 | NA | MAD1L1 Entrez, Source | MAD1 mitotic arrest deficient-like 1 (yeast) | 6685 | 0.431 | 0.4364 | No |
| 10 | CLCN3 | NA | CLCN3 Entrez, Source | chloride channel 3 | 6866 | 0.411 | 0.4474 | No |
| 11 | POU6F2 | NA | POU6F2 Entrez, Source | POU domain, class 6, transcription factor 2 | 7415 | 0.352 | 0.4423 | No |
| 12 | ADRA1A | NA | ADRA1A Entrez, Source | adrenergic, alpha-1A-, receptor | 7881 | 0.310 | 0.4385 | No |
| 13 | PRTN3 | NA | PRTN3 Entrez, Source | proteinase 3 (serine proteinase, neutrophil, Wegener granulomatosis autoantigen) | 8235 | 0.283 | 0.4376 | No |
| 14 | PRM2 | NA | PRM2 Entrez, Source | protamine 2 | 8277 | 0.279 | 0.4480 | No |
| 15 | ZFY | NA | ZFY Entrez, Source | zinc finger protein, Y-linked | 8605 | 0.256 | 0.4470 | No |
| 16 | CD79B | NA | CD79B Entrez, Source | CD79b molecule, immunoglobulin-associated beta | 9103 | 0.224 | 0.4383 | No |
| 17 | GRAP | NA | GRAP Entrez, Source | GRB2-related adaptor protein | 9307 | 0.213 | 0.4400 | No |
| 18 | SLC28A1 | NA | SLC28A1 Entrez, Source | solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 | 11272 | 0.119 | 0.3731 | No |
| 19 | GABRA2 | NA | GABRA2 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 2 | 12036 | 0.089 | 0.3490 | No |
| 20 | GDF5 | NA | GDF5 Entrez, Source | growth differentiation factor 5 (cartilage-derived morphogenetic protein-1) | 14866 | -0.016 | 0.2460 | No |
| 21 | SLURP1 | NA | SLURP1 Entrez, Source | secreted LY6/PLAUR domain containing 1 | 15581 | -0.044 | 0.2217 | No |
| 22 | HPCA | NA | HPCA Entrez, Source | hippocalcin | 15839 | -0.053 | 0.2146 | No |
| 23 | IFNA4 | NA | IFNA4 Entrez, Source | interferon, alpha 4 | 16416 | -0.078 | 0.1968 | No |
| 24 | REM1 | NA | REM1 Entrez, Source | RAS (RAD and GEM)-like GTP-binding 1 | 17075 | -0.104 | 0.1771 | No |
| 25 | VIPR2 | NA | VIPR2 Entrez, Source | vasoactive intestinal peptide receptor 2 | 18050 | -0.156 | 0.1481 | No |
| 26 | SAG | NA | SAG Entrez, Source | S-antigen; retina and pineal gland (arrestin) | 19141 | -0.238 | 0.1183 | No |
| 27 | SGCA | NA | SGCA Entrez, Source | sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) | 20289 | -0.354 | 0.0914 | No |
| 28 | SPTLC2 | NA | SPTLC2 Entrez, Source | serine palmitoyltransferase, long chain base subunit 2 | 21539 | -0.541 | 0.0687 | No |
| 29 | ITGAM | NA | ITGAM Entrez, Source | integrin, alpha M (complement component 3 receptor 3 subunit) | 22152 | -0.657 | 0.0743 | No |
| 30 | NTSR1 | NA | NTSR1 Entrez, Source | neurotensin receptor 1 (high affinity) | 24807 | -1.347 | 0.0346 | No |
| 31 | GABRA3 | NA | GABRA3 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | 24833 | -1.355 | 0.0914 | No |