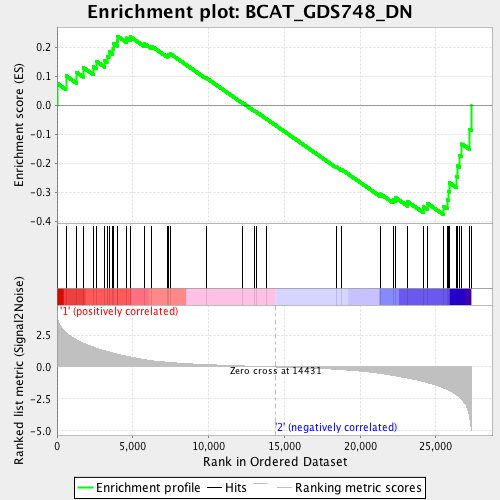
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | BCAT_GDS748_DN |
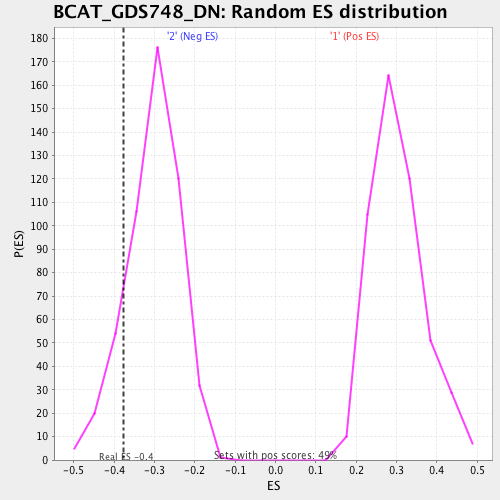
| Enrichment Score (ES) | -0.3773706 |
| Normalized Enrichment Score (NES) | -1.247063 |
| Nominal p-value | 0.14007781 |
| FDR q-value | 0.20662214 |
| FWER p-Value | 0.852 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GAS1 | NA | GAS1 Entrez, Source | growth arrest-specific 1 | 1 | 4.318 | 0.0763 | No |
| 2 | BMPR1A | NA | BMPR1A Entrez, Source | bone morphogenetic protein receptor, type IA | 587 | 2.687 | 0.1023 | No |
| 3 | TCF25 | NA | TCF25 Entrez, Source | transcription factor 25 (basic helix-loop-helix) | 1298 | 2.102 | 0.1135 | No |
| 4 | ACAD8 | NA | ACAD8 Entrez, Source | acyl-Coenzyme A dehydrogenase family, member 8 | 1720 | 1.863 | 0.1310 | No |
| 5 | FAM46A | NA | FAM46A Entrez, Source | family with sequence similarity 46, member A | 2390 | 1.553 | 0.1339 | No |
| 6 | MLH1 | NA | MLH1 Entrez, Source | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | 2579 | 1.470 | 0.1530 | No |
| 7 | SNAI2 | NA | SNAI2 Entrez, Source | snail homolog 2 (Drosophila) | 3138 | 1.262 | 0.1548 | No |
| 8 | SLC35E3 | NA | SLC35E3 Entrez, Source | solute carrier family 35, member E3 | 3294 | 1.216 | 0.1706 | No |
| 9 | VPS13B | NA | VPS13B Entrez, Source | vacuolar protein sorting 13B (yeast) | 3427 | 1.175 | 0.1866 | No |
| 10 | ZNF273 | NA | ZNF273 Entrez, Source | zinc finger protein 273 | 3686 | 1.086 | 0.1963 | No |
| 11 | ZBED5 | NA | ZBED5 Entrez, Source | zinc finger, BED-type containing 5 | 3741 | 1.071 | 0.2132 | No |
| 12 | CELSR1 | NA | CELSR1 Entrez, Source | cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) | 3960 | 0.998 | 0.2229 | No |
| 13 | CDK5RAP3 | NA | CDK5RAP3 Entrez, Source | CDK5 regulatory subunit associated protein 3 | 3982 | 0.991 | 0.2396 | No |
| 14 | UROS | NA | UROS Entrez, Source | uroporphyrinogen III synthase (congenital erythropoietic porphyria) | 4567 | 0.847 | 0.2332 | No |
| 15 | ARL6IP1 | NA | ARL6IP1 Entrez, Source | ADP-ribosylation factor-like 6 interacting protein 1 | 4814 | 0.785 | 0.2381 | No |
| 16 | EXOC2 | NA | EXOC2 Entrez, Source | exocyst complex component 2 | 5766 | 0.567 | 0.2132 | No |
| 17 | CHMP4A | NA | CHMP4A Entrez, Source | chromatin modifying protein 4A | 6258 | 0.488 | 0.2039 | No |
| 18 | C5orf42 | NA | 7257 | 0.370 | 0.1738 | No | ||
| 19 | TTR | NA | TTR Entrez, Source | transthyretin (prealbumin, amyloidosis type I) | 7362 | 0.359 | 0.1763 | No |
| 20 | COL2A1 | NA | COL2A1 Entrez, Source | collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 7453 | 0.349 | 0.1792 | No |
| 21 | RALY | NA | RALY Entrez, Source | RNA binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) | 9840 | 0.186 | 0.0951 | No |
| 22 | RPS6KA5 | NA | RPS6KA5 Entrez, Source | ribosomal protein S6 kinase, 90kDa, polypeptide 5 | 12256 | 0.081 | 0.0080 | No |
| 23 | DDIT4 | NA | DDIT4 Entrez, Source | DNA-damage-inducible transcript 4 | 13010 | 0.053 | -0.0187 | No |
| 24 | HSDL2 | NA | HSDL2 Entrez, Source | hydroxysteroid dehydrogenase like 2 | 13123 | 0.048 | -0.0219 | No |
| 25 | PBLD | NA | 13798 | 0.022 | -0.0462 | No | ||
| 26 | ADAMTS20 | NA | ADAMTS20 Entrez, Source | ADAM metallopeptidase with thrombospondin type 1 motif, 20 | 18404 | -0.178 | -0.2118 | No |
| 27 | LOC654056 | NA | LOC654056 Entrez, Source | - | 18774 | -0.208 | -0.2217 | No |
| 28 | HMG20B | NA | HMG20B Entrez, Source | high-mobility group 20B | 21329 | -0.502 | -0.3064 | No |
| 29 | RDH5 | NA | RDH5 Entrez, Source | retinol dehydrogenase 5 (11-cis/9-cis) | 22170 | -0.660 | -0.3255 | No |
| 30 | PMFBP1 | NA | PMFBP1 Entrez, Source | polyamine modulated factor 1 binding protein 1 | 22349 | -0.696 | -0.3198 | No |
| 31 | SKP2 | NA | SKP2 Entrez, Source | S-phase kinase-associated protein 2 (p45) | 23129 | -0.864 | -0.3330 | No |
| 32 | ZNF32 | NA | ZNF32 Entrez, Source | zinc finger protein 32 | 24158 | -1.138 | -0.3506 | Yes |
| 33 | PABPC3 | NA | PABPC3 Entrez, Source | poly(A) binding protein, cytoplasmic 3 | 24445 | -1.225 | -0.3394 | Yes |
| 34 | ISYNA1 | NA | ISYNA1 Entrez, Source | - | 25481 | -1.631 | -0.3485 | Yes |
| 35 | MYBL1 | NA | MYBL1 Entrez, Source | v-myb myeloblastosis viral oncogene homolog (avian)-like 1 | 25746 | -1.764 | -0.3270 | Yes |
| 36 | LAP3 | NA | LAP3 Entrez, Source | leucine aminopeptidase 3 | 25812 | -1.803 | -0.2975 | Yes |
| 37 | PDHA1 | NA | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 25871 | -1.839 | -0.2672 | Yes |
| 38 | CNPY3 | NA | 26363 | -2.182 | -0.2466 | Yes | ||
| 39 | HLX | NA | 26424 | -2.232 | -0.2094 | Yes | ||
| 40 | ZNF318 | NA | ZNF318 Entrez, Source | zinc finger protein 318 | 26574 | -2.359 | -0.1731 | Yes |
| 41 | SLC27A6 | NA | SLC27A6 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 6 | 26703 | -2.509 | -0.1335 | Yes |
| 42 | BASP1 | NA | BASP1 Entrez, Source | brain abundant, membrane attached signal protein 1 | 27223 | -3.935 | -0.0829 | Yes |
| 43 | KLHL9 | NA | KLHL9 Entrez, Source | kelch-like 9 (Drosophila) | 27327 | -4.911 | 0.0001 | Yes |