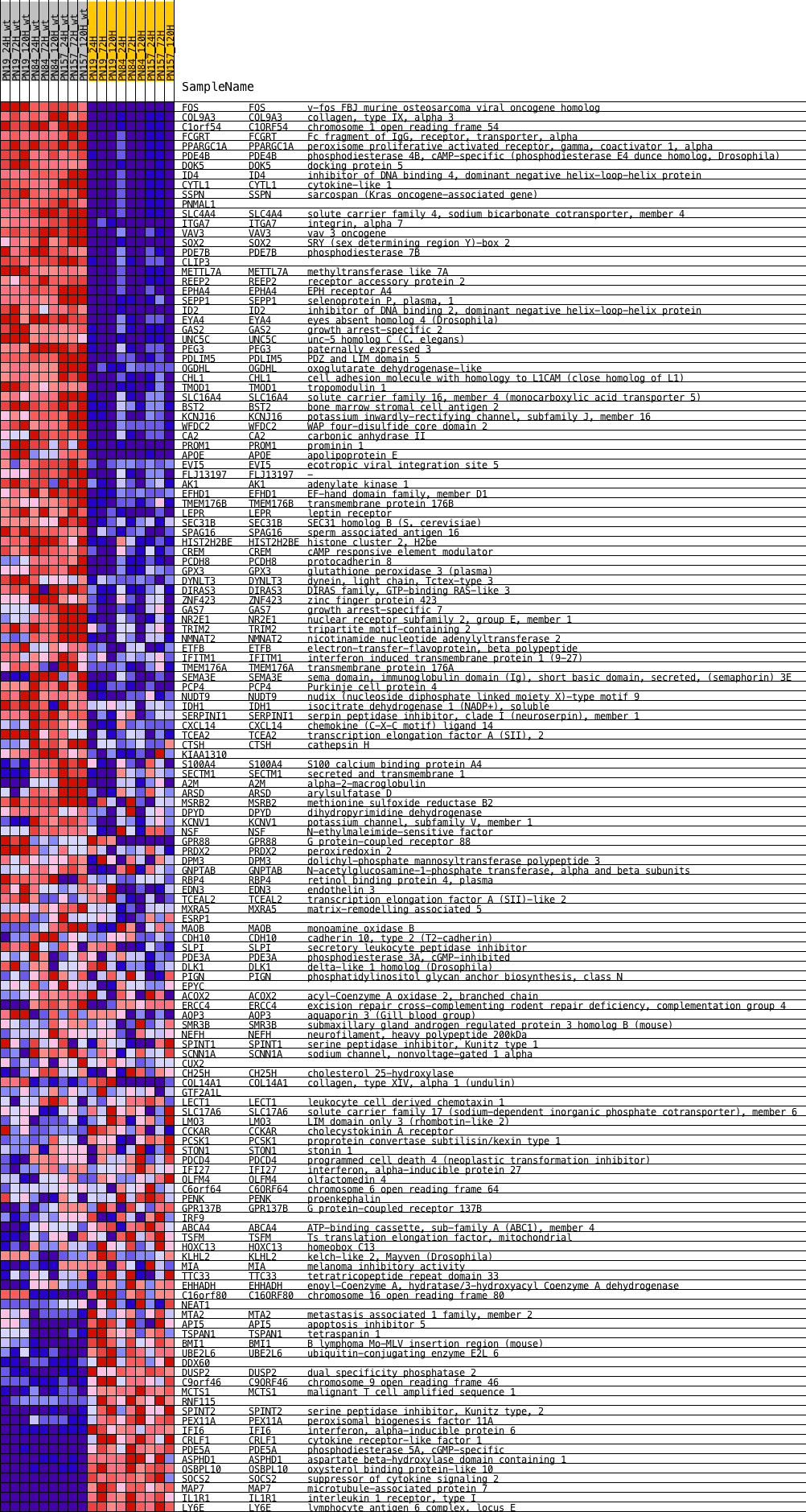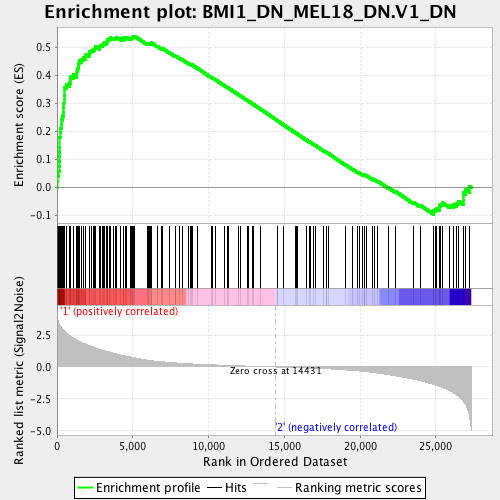
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | BMI1_DN_MEL18_DN.V1_DN |
| Enrichment Score (ES) | 0.53882086 |
| Normalized Enrichment Score (NES) | 1.6225693 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0109116165 |
| FWER p-Value | 0.077 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FOS | NA | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 3 | 4.177 | 0.0221 | Yes |
| 2 | COL9A3 | NA | COL9A3 Entrez, Source | collagen, type IX, alpha 3 | 38 | 3.730 | 0.0406 | Yes |
| 3 | C1orf54 | NA | C1ORF54 Entrez, Source | chromosome 1 open reading frame 54 | 85 | 3.528 | 0.0577 | Yes |
| 4 | FCGRT | NA | FCGRT Entrez, Source | Fc fragment of IgG, receptor, transporter, alpha | 131 | 3.400 | 0.0740 | Yes |
| 5 | PPARGC1A | NA | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 136 | 3.387 | 0.0919 | Yes |
| 6 | PDE4B | NA | PDE4B Entrez, Source | phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | 147 | 3.365 | 0.1094 | Yes |
| 7 | DOK5 | NA | DOK5 Entrez, Source | docking protein 5 | 153 | 3.346 | 0.1270 | Yes |
| 8 | ID4 | NA | ID4 Entrez, Source | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | 172 | 3.304 | 0.1438 | Yes |
| 9 | CYTL1 | NA | CYTL1 Entrez, Source | cytokine-like 1 | 179 | 3.299 | 0.1611 | Yes |
| 10 | SSPN | NA | SSPN Entrez, Source | sarcospan (Kras oncogene-associated gene) | 180 | 3.298 | 0.1786 | Yes |
| 11 | PNMAL1 | NA | 200 | 3.272 | 0.1953 | Yes | ||
| 12 | SLC4A4 | NA | SLC4A4 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 4 | 223 | 3.234 | 0.2117 | Yes |
| 13 | ITGA7 | NA | ITGA7 Entrez, Source | integrin, alpha 7 | 307 | 3.066 | 0.2249 | Yes |
| 14 | VAV3 | NA | VAV3 Entrez, Source | vav 3 oncogene | 319 | 3.047 | 0.2407 | Yes |
| 15 | SOX2 | NA | SOX2 Entrez, Source | SRY (sex determining region Y)-box 2 | 342 | 3.009 | 0.2558 | Yes |
| 16 | PDE7B | NA | PDE7B Entrez, Source | phosphodiesterase 7B | 411 | 2.910 | 0.2688 | Yes |
| 17 | CLIP3 | NA | 421 | 2.902 | 0.2839 | Yes | ||
| 18 | METTL7A | NA | METTL7A Entrez, Source | methyltransferase like 7A | 433 | 2.889 | 0.2988 | Yes |
| 19 | REEP2 | NA | REEP2 Entrez, Source | receptor accessory protein 2 | 455 | 2.863 | 0.3132 | Yes |
| 20 | EPHA4 | NA | EPHA4 Entrez, Source | EPH receptor A4 | 459 | 2.861 | 0.3283 | Yes |
| 21 | SEPP1 | NA | SEPP1 Entrez, Source | selenoprotein P, plasma, 1 | 479 | 2.830 | 0.3426 | Yes |
| 22 | ID2 | NA | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 481 | 2.829 | 0.3576 | Yes |
| 23 | EYA4 | NA | EYA4 Entrez, Source | eyes absent homolog 4 (Drosophila) | 612 | 2.664 | 0.3670 | Yes |
| 24 | GAS2 | NA | GAS2 Entrez, Source | growth arrest-specific 2 | 810 | 2.465 | 0.3728 | Yes |
| 25 | UNC5C | NA | UNC5C Entrez, Source | unc-5 homolog C (C. elegans) | 878 | 2.408 | 0.3831 | Yes |
| 26 | PEG3 | NA | PEG3 Entrez, Source | paternally expressed 3 | 886 | 2.400 | 0.3956 | Yes |
| 27 | PDLIM5 | NA | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 1063 | 2.270 | 0.4012 | Yes |
| 28 | OGDHL | NA | OGDHL Entrez, Source | oxoglutarate dehydrogenase-like | 1296 | 2.103 | 0.4038 | Yes |
| 29 | CHL1 | NA | CHL1 Entrez, Source | cell adhesion molecule with homology to L1CAM (close homolog of L1) | 1309 | 2.095 | 0.4145 | Yes |
| 30 | TMOD1 | NA | TMOD1 Entrez, Source | tropomodulin 1 | 1335 | 2.081 | 0.4246 | Yes |
| 31 | SLC16A4 | NA | SLC16A4 Entrez, Source | solute carrier family 16, member 4 (monocarboxylic acid transporter 5) | 1412 | 2.026 | 0.4326 | Yes |
| 32 | BST2 | NA | BST2 Entrez, Source | bone marrow stromal cell antigen 2 | 1430 | 2.017 | 0.4427 | Yes |
| 33 | KCNJ16 | NA | KCNJ16 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 16 | 1471 | 1.995 | 0.4518 | Yes |
| 34 | WFDC2 | NA | WFDC2 Entrez, Source | WAP four-disulfide core domain 2 | 1637 | 1.902 | 0.4558 | Yes |
| 35 | CA2 | NA | CA2 Entrez, Source | carbonic anhydrase II | 1735 | 1.854 | 0.4621 | Yes |
| 36 | PROM1 | NA | PROM1 Entrez, Source | prominin 1 | 1895 | 1.778 | 0.4657 | Yes |
| 37 | APOE | NA | APOE Entrez, Source | apolipoprotein E | 1901 | 1.776 | 0.4749 | Yes |
| 38 | EVI5 | NA | EVI5 Entrez, Source | ecotropic viral integration site 5 | 2106 | 1.675 | 0.4763 | Yes |
| 39 | FLJ13197 | NA | FLJ13197 Entrez, Source | - | 2136 | 1.662 | 0.4841 | Yes |
| 40 | AK1 | NA | AK1 Entrez, Source | adenylate kinase 1 | 2256 | 1.615 | 0.4883 | Yes |
| 41 | EFHD1 | NA | EFHD1 Entrez, Source | EF-hand domain family, member D1 | 2376 | 1.561 | 0.4922 | Yes |
| 42 | TMEM176B | NA | TMEM176B Entrez, Source | transmembrane protein 176B | 2498 | 1.506 | 0.4957 | Yes |
| 43 | LEPR | NA | LEPR Entrez, Source | leptin receptor | 2538 | 1.487 | 0.5022 | Yes |
| 44 | SEC31B | NA | SEC31B Entrez, Source | SEC31 homolog B (S. cerevisiae) | 2805 | 1.385 | 0.4997 | Yes |
| 45 | SPAG16 | NA | SPAG16 Entrez, Source | sperm associated antigen 16 | 2830 | 1.375 | 0.5062 | Yes |
| 46 | HIST2H2BE | NA | HIST2H2BE Entrez, Source | histone cluster 2, H2be | 2976 | 1.318 | 0.5078 | Yes |
| 47 | CREM | NA | CREM Entrez, Source | cAMP responsive element modulator | 3043 | 1.296 | 0.5123 | Yes |
| 48 | PCDH8 | NA | PCDH8 Entrez, Source | protocadherin 8 | 3119 | 1.269 | 0.5163 | Yes |
| 49 | GPX3 | NA | GPX3 Entrez, Source | glutathione peroxidase 3 (plasma) | 3278 | 1.220 | 0.5169 | Yes |
| 50 | DYNLT3 | NA | DYNLT3 Entrez, Source | dynein, light chain, Tctex-type 3 | 3297 | 1.215 | 0.5227 | Yes |
| 51 | DIRAS3 | NA | DIRAS3 Entrez, Source | DIRAS family, GTP-binding RAS-like 3 | 3323 | 1.208 | 0.5282 | Yes |
| 52 | ZNF423 | NA | ZNF423 Entrez, Source | zinc finger protein 423 | 3454 | 1.167 | 0.5296 | Yes |
| 53 | GAS7 | NA | GAS7 Entrez, Source | growth arrest-specific 7 | 3520 | 1.148 | 0.5333 | Yes |
| 54 | NR2E1 | NA | NR2E1 Entrez, Source | nuclear receptor subfamily 2, group E, member 1 | 3718 | 1.076 | 0.5318 | Yes |
| 55 | TRIM2 | NA | TRIM2 Entrez, Source | tripartite motif-containing 2 | 3879 | 1.024 | 0.5313 | Yes |
| 56 | NMNAT2 | NA | NMNAT2 Entrez, Source | nicotinamide nucleotide adenylyltransferase 2 | 3931 | 1.008 | 0.5348 | Yes |
| 57 | ETFB | NA | ETFB Entrez, Source | electron-transfer-flavoprotein, beta polypeptide | 4202 | 0.931 | 0.5298 | Yes |
| 58 | IFITM1 | NA | IFITM1 Entrez, Source | interferon induced transmembrane protein 1 (9-27) | 4366 | 0.891 | 0.5286 | Yes |
| 59 | TMEM176A | NA | TMEM176A Entrez, Source | transmembrane protein 176A | 4393 | 0.884 | 0.5323 | Yes |
| 60 | SEMA3E | NA | SEMA3E Entrez, Source | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E | 4517 | 0.857 | 0.5323 | Yes |
| 61 | PCP4 | NA | PCP4 Entrez, Source | Purkinje cell protein 4 | 4601 | 0.842 | 0.5337 | Yes |
| 62 | NUDT9 | NA | NUDT9 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 9 | 4838 | 0.779 | 0.5292 | Yes |
| 63 | IDH1 | NA | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 4940 | 0.757 | 0.5295 | Yes |
| 64 | SERPINI1 | NA | SERPINI1 Entrez, Source | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 4947 | 0.755 | 0.5333 | Yes |
| 65 | CXCL14 | NA | CXCL14 Entrez, Source | chemokine (C-X-C motif) ligand 14 | 4987 | 0.746 | 0.5358 | Yes |
| 66 | TCEA2 | NA | TCEA2 Entrez, Source | transcription elongation factor A (SII), 2 | 5013 | 0.740 | 0.5388 | Yes |
| 67 | CTSH | NA | CTSH Entrez, Source | cathepsin H | 5119 | 0.715 | 0.5388 | No |
| 68 | KIAA1310 | NA | 5950 | 0.537 | 0.5111 | No | ||
| 69 | S100A4 | NA | S100A4 Entrez, Source | S100 calcium binding protein A4 | 6035 | 0.521 | 0.5107 | No |
| 70 | SECTM1 | NA | SECTM1 Entrez, Source | secreted and transmembrane 1 | 6078 | 0.515 | 0.5119 | No |
| 71 | A2M | NA | A2M Entrez, Source | alpha-2-macroglobulin | 6145 | 0.506 | 0.5122 | No |
| 72 | ARSD | NA | ARSD Entrez, Source | arylsulfatase D | 6195 | 0.497 | 0.5130 | No |
| 73 | MSRB2 | NA | MSRB2 Entrez, Source | methionine sulfoxide reductase B2 | 6203 | 0.497 | 0.5154 | No |
| 74 | DPYD | NA | DPYD Entrez, Source | dihydropyrimidine dehydrogenase | 6599 | 0.441 | 0.5032 | No |
| 75 | KCNV1 | NA | KCNV1 Entrez, Source | potassium channel, subfamily V, member 1 | 6890 | 0.409 | 0.4947 | No |
| 76 | NSF | NA | NSF Entrez, Source | N-ethylmaleimide-sensitive factor | 6960 | 0.401 | 0.4943 | No |
| 77 | GPR88 | NA | GPR88 Entrez, Source | G protein-coupled receptor 88 | 7382 | 0.357 | 0.4807 | No |
| 78 | PRDX2 | NA | PRDX2 Entrez, Source | peroxiredoxin 2 | 7779 | 0.320 | 0.4679 | No |
| 79 | DPM3 | NA | DPM3 Entrez, Source | dolichyl-phosphate mannosyltransferase polypeptide 3 | 7834 | 0.315 | 0.4675 | No |
| 80 | GNPTAB | NA | GNPTAB Entrez, Source | N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits | 8077 | 0.294 | 0.4602 | No |
| 81 | RBP4 | NA | RBP4 Entrez, Source | retinol binding protein 4, plasma | 8284 | 0.278 | 0.4541 | No |
| 82 | EDN3 | NA | EDN3 Entrez, Source | endothelin 3 | 8684 | 0.250 | 0.4407 | No |
| 83 | TCEAL2 | NA | TCEAL2 Entrez, Source | transcription elongation factor A (SII)-like 2 | 8803 | 0.241 | 0.4377 | No |
| 84 | MXRA5 | NA | MXRA5 Entrez, Source | matrix-remodelling associated 5 | 8842 | 0.239 | 0.4376 | No |
| 85 | ESRP1 | NA | 8900 | 0.236 | 0.4367 | No | ||
| 86 | MAOB | NA | MAOB Entrez, Source | monoamine oxidase B | 9285 | 0.215 | 0.4237 | No |
| 87 | CDH10 | NA | CDH10 Entrez, Source | cadherin 10, type 2 (T2-cadherin) | 10163 | 0.172 | 0.3924 | No |
| 88 | SLPI | NA | SLPI Entrez, Source | secretory leukocyte peptidase inhibitor | 10226 | 0.168 | 0.3910 | No |
| 89 | PDE3A | NA | PDE3A Entrez, Source | phosphodiesterase 3A, cGMP-inhibited | 10425 | 0.159 | 0.3845 | No |
| 90 | DLK1 | NA | DLK1 Entrez, Source | delta-like 1 homolog (Drosophila) | 11014 | 0.132 | 0.3636 | No |
| 91 | PIGN | NA | PIGN Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class N | 11217 | 0.122 | 0.3568 | No |
| 92 | EPYC | NA | 11294 | 0.118 | 0.3547 | No | ||
| 93 | ACOX2 | NA | ACOX2 Entrez, Source | acyl-Coenzyme A oxidase 2, branched chain | 11947 | 0.092 | 0.3312 | No |
| 94 | ERCC4 | NA | ERCC4 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 4 | 12069 | 0.088 | 0.3272 | No |
| 95 | AQP3 | NA | AQP3 Entrez, Source | aquaporin 3 (Gill blood group) | 12539 | 0.070 | 0.3103 | No |
| 96 | SMR3B | NA | SMR3B Entrez, Source | submaxillary gland androgen regulated protein 3 homolog B (mouse) | 12625 | 0.066 | 0.3075 | No |
| 97 | NEFH | NA | NEFH Entrez, Source | neurofilament, heavy polypeptide 200kDa | 12921 | 0.056 | 0.2970 | No |
| 98 | SPINT1 | NA | SPINT1 Entrez, Source | serine peptidase inhibitor, Kunitz type 1 | 12941 | 0.055 | 0.2966 | No |
| 99 | SCNN1A | NA | SCNN1A Entrez, Source | sodium channel, nonvoltage-gated 1 alpha | 13451 | 0.035 | 0.2780 | No |
| 100 | CUX2 | NA | 14555 | -0.004 | 0.2375 | No | ||
| 101 | CH25H | NA | CH25H Entrez, Source | cholesterol 25-hydroxylase | 14556 | -0.004 | 0.2375 | No |
| 102 | COL14A1 | NA | COL14A1 Entrez, Source | collagen, type XIV, alpha 1 (undulin) | 14969 | -0.020 | 0.2224 | No |
| 103 | GTF2A1L | NA | 15724 | -0.049 | 0.1950 | No | ||
| 104 | LECT1 | NA | LECT1 Entrez, Source | leukocyte cell derived chemotaxin 1 | 15777 | -0.051 | 0.1933 | No |
| 105 | SLC17A6 | NA | SLC17A6 Entrez, Source | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 | 15850 | -0.054 | 0.1910 | No |
| 106 | LMO3 | NA | LMO3 Entrez, Source | LIM domain only 3 (rhombotin-like 2) | 16466 | -0.080 | 0.1688 | No |
| 107 | CCKAR | NA | CCKAR Entrez, Source | cholecystokinin A receptor | 16682 | -0.088 | 0.1613 | No |
| 108 | PCSK1 | NA | PCSK1 Entrez, Source | proprotein convertase subtilisin/kexin type 1 | 16703 | -0.089 | 0.1611 | No |
| 109 | STON1 | NA | STON1 Entrez, Source | stonin 1 | 16900 | -0.097 | 0.1544 | No |
| 110 | PDCD4 | NA | PDCD4 Entrez, Source | programmed cell death 4 (neoplastic transformation inhibitor) | 17060 | -0.104 | 0.1491 | No |
| 111 | IFI27 | NA | IFI27 Entrez, Source | interferon, alpha-inducible protein 27 | 17553 | -0.127 | 0.1316 | No |
| 112 | OLFM4 | NA | OLFM4 Entrez, Source | olfactomedin 4 | 17764 | -0.138 | 0.1247 | No |
| 113 | C6orf64 | NA | C6ORF64 Entrez, Source | chromosome 6 open reading frame 64 | 17783 | -0.139 | 0.1247 | No |
| 114 | PENK | NA | PENK Entrez, Source | proenkephalin | 17908 | -0.148 | 0.1210 | No |
| 115 | GPR137B | NA | GPR137B Entrez, Source | G protein-coupled receptor 137B | 19045 | -0.231 | 0.0804 | No |
| 116 | IRF9 | NA | 19523 | -0.272 | 0.0643 | No | ||
| 117 | ABCA4 | NA | ABCA4 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 4 | 19843 | -0.304 | 0.0542 | No |
| 118 | TSFM | NA | TSFM Entrez, Source | Ts translation elongation factor, mitochondrial | 19929 | -0.313 | 0.0527 | No |
| 119 | HOXC13 | NA | HOXC13 Entrez, Source | homeobox C13 | 20171 | -0.339 | 0.0456 | No |
| 120 | KLHL2 | NA | KLHL2 Entrez, Source | kelch-like 2, Mayven (Drosophila) | 20265 | -0.350 | 0.0441 | No |
| 121 | MIA | NA | MIA Entrez, Source | melanoma inhibitory activity | 20282 | -0.352 | 0.0454 | No |
| 122 | TTC33 | NA | TTC33 Entrez, Source | tetratricopeptide repeat domain 33 | 20419 | -0.370 | 0.0423 | No |
| 123 | EHHADH | NA | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 20792 | -0.421 | 0.0309 | No |
| 124 | C16orf80 | NA | C16ORF80 Entrez, Source | chromosome 16 open reading frame 80 | 20949 | -0.445 | 0.0275 | No |
| 125 | NEAT1 | NA | 21149 | -0.474 | 0.0227 | No | ||
| 126 | MTA2 | NA | MTA2 Entrez, Source | metastasis associated 1 family, member 2 | 21849 | -0.599 | 0.0002 | No |
| 127 | API5 | NA | API5 Entrez, Source | apoptosis inhibitor 5 | 22332 | -0.694 | -0.0139 | No |
| 128 | TSPAN1 | NA | TSPAN1 Entrez, Source | tetraspanin 1 | 23502 | -0.951 | -0.0518 | No |
| 129 | BMI1 | NA | BMI1 Entrez, Source | B lymphoma Mo-MLV insertion region (mouse) | 23952 | -1.077 | -0.0626 | No |
| 130 | UBE2L6 | NA | UBE2L6 Entrez, Source | ubiquitin-conjugating enzyme E2L 6 | 24851 | -1.360 | -0.0885 | No |
| 131 | DDX60 | NA | 24864 | -1.366 | -0.0816 | No | ||
| 132 | DUSP2 | NA | DUSP2 Entrez, Source | dual specificity phosphatase 2 | 24949 | -1.396 | -0.0773 | No |
| 133 | C9orf46 | NA | C9ORF46 Entrez, Source | chromosome 9 open reading frame 46 | 25030 | -1.431 | -0.0727 | No |
| 134 | MCTS1 | NA | MCTS1 Entrez, Source | malignant T cell amplified sequence 1 | 25232 | -1.514 | -0.0720 | No |
| 135 | RNF115 | NA | 25236 | -1.517 | -0.0641 | No | ||
| 136 | SPINT2 | NA | SPINT2 Entrez, Source | serine peptidase inhibitor, Kunitz type, 2 | 25309 | -1.552 | -0.0585 | No |
| 137 | PEX11A | NA | PEX11A Entrez, Source | peroxisomal biogenesis factor 11A | 25429 | -1.608 | -0.0543 | No |
| 138 | IFI6 | NA | IFI6 Entrez, Source | interferon, alpha-inducible protein 6 | 25912 | -1.862 | -0.0622 | No |
| 139 | CRLF1 | NA | CRLF1 Entrez, Source | cytokine receptor-like factor 1 | 26139 | -2.008 | -0.0598 | No |
| 140 | PDE5A | NA | PDE5A Entrez, Source | phosphodiesterase 5A, cGMP-specific | 26369 | -2.191 | -0.0566 | No |
| 141 | ASPHD1 | NA | ASPHD1 Entrez, Source | aspartate beta-hydroxylase domain containing 1 | 26501 | -2.302 | -0.0492 | No |
| 142 | OSBPL10 | NA | OSBPL10 Entrez, Source | oxysterol binding protein-like 10 | 26811 | -2.666 | -0.0464 | No |
| 143 | SOCS2 | NA | SOCS2 Entrez, Source | suppressor of cytokine signaling 2 | 26833 | -2.703 | -0.0328 | No |
| 144 | MAP7 | NA | MAP7 Entrez, Source | microtubule-associated protein 7 | 26837 | -2.718 | -0.0185 | No |
| 145 | IL1R1 | NA | IL1R1 Entrez, Source | interleukin 1 receptor, type I | 26964 | -2.933 | -0.0076 | No |
| 146 | LY6E | NA | LY6E Entrez, Source | lymphocyte antigen 6 complex, locus E | 27224 | -3.952 | 0.0039 | No |