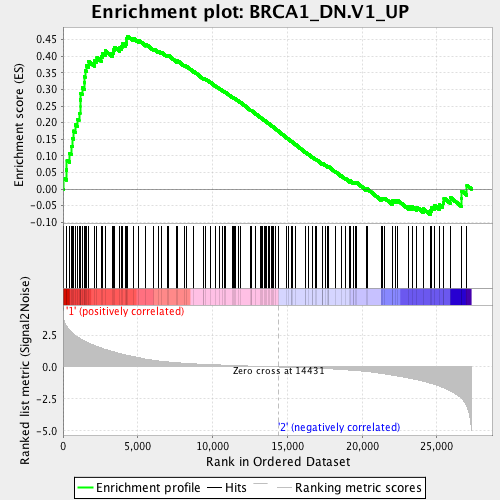
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | BRCA1_DN.V1_UP |
| Enrichment Score (ES) | 0.46068618 |
| Normalized Enrichment Score (NES) | 1.8761055 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0024836194 |
| FWER p-Value | 0.003 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAP2 | NA | MAP2 Entrez, Source | microtubule-associated protein 2 | 54 | 3.643 | 0.0330 | Yes |
| 2 | RNF122 | NA | RNF122 Entrez, Source | ring finger protein 122 | 213 | 3.247 | 0.0583 | Yes |
| 3 | TNC | NA | TNC Entrez, Source | tenascin C (hexabrachion) | 258 | 3.160 | 0.0870 | Yes |
| 4 | ZNF529 | NA | ZNF529 Entrez, Source | zinc finger protein 529 | 436 | 2.888 | 0.1082 | Yes |
| 5 | KCNA2 | NA | KCNA2 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 2 | 563 | 2.703 | 0.1295 | Yes |
| 6 | ZNF682 | NA | ZNF682 Entrez, Source | zinc finger protein 682 | 638 | 2.636 | 0.1521 | Yes |
| 7 | LEPREL2 | NA | LEPREL2 Entrez, Source | leprecan-like 2 | 691 | 2.587 | 0.1750 | Yes |
| 8 | CYP2U1 | NA | CYP2U1 Entrez, Source | cytochrome P450, family 2, subfamily U, polypeptide 1 | 831 | 2.447 | 0.1934 | Yes |
| 9 | CTNNA3 | NA | CTNNA3 Entrez, Source | catenin (cadherin-associated protein), alpha 3 | 982 | 2.326 | 0.2102 | Yes |
| 10 | LHX6 | NA | LHX6 Entrez, Source | LIM homeobox 6 | 1065 | 2.269 | 0.2290 | Yes |
| 11 | THBS2 | NA | THBS2 Entrez, Source | thrombospondin 2 | 1132 | 2.224 | 0.2479 | Yes |
| 12 | LDB2 | NA | LDB2 Entrez, Source | LIM domain binding 2 | 1134 | 2.223 | 0.2692 | Yes |
| 13 | TIMP4 | NA | TIMP4 Entrez, Source | TIMP metallopeptidase inhibitor 4 | 1166 | 2.198 | 0.2891 | Yes |
| 14 | GABRB1 | NA | GABRB1 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, beta 1 | 1274 | 2.119 | 0.3055 | Yes |
| 15 | SLC16A4 | NA | SLC16A4 Entrez, Source | solute carrier family 16, member 4 (monocarboxylic acid transporter 5) | 1412 | 2.026 | 0.3199 | Yes |
| 16 | STAR | NA | STAR Entrez, Source | steroidogenic acute regulator | 1425 | 2.019 | 0.3389 | Yes |
| 17 | KCNJ16 | NA | KCNJ16 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 16 | 1471 | 1.995 | 0.3564 | Yes |
| 18 | NKX2-2 | NA | NKX2-2 Entrez, Source | NK2 transcription factor related, locus 2 (Drosophila) | 1562 | 1.945 | 0.3717 | Yes |
| 19 | PTPRS | NA | PTPRS Entrez, Source | protein tyrosine phosphatase, receptor type, S | 1709 | 1.867 | 0.3843 | Yes |
| 20 | CNTN6 | NA | CNTN6 Entrez, Source | contactin 6 | 2089 | 1.682 | 0.3865 | Yes |
| 21 | CILP | NA | CILP Entrez, Source | cartilage intermediate layer protein, nucleotide pyrophosphohydrolase | 2262 | 1.612 | 0.3956 | Yes |
| 22 | IPW | NA | IPW Entrez, Source | imprinted in Prader-Willi syndrome | 2573 | 1.471 | 0.3983 | Yes |
| 23 | APBB1 | NA | APBB1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | 2642 | 1.447 | 0.4097 | Yes |
| 24 | FAM46C | NA | FAM46C Entrez, Source | family with sequence similarity 46, member C | 2818 | 1.380 | 0.4165 | Yes |
| 25 | TTLL7 | NA | TTLL7 Entrez, Source | tubulin tyrosine ligase-like family, member 7 | 3317 | 1.209 | 0.4098 | Yes |
| 26 | LRRTM2 | NA | LRRTM2 Entrez, Source | leucine rich repeat transmembrane neuronal 2 | 3374 | 1.192 | 0.4192 | Yes |
| 27 | ZNF423 | NA | ZNF423 Entrez, Source | zinc finger protein 423 | 3454 | 1.167 | 0.4275 | Yes |
| 28 | FCGR2C | NA | FCGR2C Entrez, Source | Fc fragment of IgG, low affinity IIb, receptor for (CD32) | 3794 | 1.053 | 0.4251 | Yes |
| 29 | SIX1 | NA | SIX1 Entrez, Source | sine oculis homeobox homolog 1 (Drosophila) | 3916 | 1.012 | 0.4304 | Yes |
| 30 | CG030 | NA | CG030 Entrez, Source | - | 3989 | 0.989 | 0.4373 | Yes |
| 31 | SP4 | NA | SP4 Entrez, Source | Sp4 transcription factor | 4172 | 0.940 | 0.4396 | Yes |
| 32 | EXTL1 | NA | EXTL1 Entrez, Source | exostoses (multiple)-like 1 | 4260 | 0.918 | 0.4452 | Yes |
| 33 | TRPM8 | NA | TRPM8 Entrez, Source | transient receptor potential cation channel, subfamily M, member 8 | 4264 | 0.917 | 0.4539 | Yes |
| 34 | B3GALT2 | NA | B3GALT2 Entrez, Source | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 | 4316 | 0.904 | 0.4607 | Yes |
| 35 | NLGN4Y | NA | NLGN4Y Entrez, Source | neuroligin 4, Y-linked | 4685 | 0.817 | 0.4550 | No |
| 36 | GRID2 | NA | GRID2 Entrez, Source | glutamate receptor, ionotropic, delta 2 | 5054 | 0.732 | 0.4485 | No |
| 37 | CD3D | NA | CD3D Entrez, Source | CD3d molecule, delta (CD3-TCR complex) | 5535 | 0.610 | 0.4367 | No |
| 38 | NPHS2 | NA | NPHS2 Entrez, Source | nephrosis 2, idiopathic, steroid-resistant (podocin) | 6079 | 0.515 | 0.4217 | No |
| 39 | WDR25 | NA | WDR25 Entrez, Source | WD repeat domain 25 | 6387 | 0.469 | 0.4149 | No |
| 40 | HIST1H2BJ | NA | HIST1H2BJ Entrez, Source | histone cluster 1, H2bj | 6566 | 0.445 | 0.4126 | No |
| 41 | GPR6 | NA | GPR6 Entrez, Source | G protein-coupled receptor 6 | 6969 | 0.400 | 0.4017 | No |
| 42 | LTC4S | NA | LTC4S Entrez, Source | leukotriene C4 synthase | 7062 | 0.391 | 0.4020 | No |
| 43 | CHAT | NA | CHAT Entrez, Source | choline acetyltransferase | 7617 | 0.334 | 0.3849 | No |
| 44 | TSSK2 | NA | TSSK2 Entrez, Source | testis-specific serine kinase 2 | 7619 | 0.334 | 0.3880 | No |
| 45 | LRMP | NA | LRMP Entrez, Source | lymphoid-restricted membrane protein | 8098 | 0.292 | 0.3733 | No |
| 46 | ANGPT4 | NA | ANGPT4 Entrez, Source | angiopoietin 4 | 8255 | 0.281 | 0.3702 | No |
| 47 | DAGLA | NA | 8756 | 0.244 | 0.3542 | No | ||
| 48 | PRSS21 | NA | PRSS21 Entrez, Source | protease, serine, 21 (testisin) | 9384 | 0.209 | 0.3331 | No |
| 49 | CCKBR | NA | CCKBR Entrez, Source | cholecystokinin B receptor | 9421 | 0.207 | 0.3338 | No |
| 50 | CD160 | NA | CD160 Entrez, Source | CD160 molecule | 9548 | 0.201 | 0.3311 | No |
| 51 | LILRP2 | NA | LILRP2 Entrez, Source | leukocyte immunoglobulin-like receptor pseudogene 2 | 9852 | 0.186 | 0.3217 | No |
| 52 | IL1RL2 | NA | IL1RL2 Entrez, Source | interleukin 1 receptor-like 2 | 10200 | 0.170 | 0.3106 | No |
| 53 | NEUROD6 | NA | NEUROD6 Entrez, Source | neurogenic differentiation 6 | 10434 | 0.159 | 0.3036 | No |
| 54 | POU2F3 | NA | POU2F3 Entrez, Source | POU domain, class 2, transcription factor 3 | 10659 | 0.149 | 0.2968 | No |
| 55 | PRAMEF12 | NA | PRAMEF12 Entrez, Source | PRAME family member 12 | 10797 | 0.141 | 0.2931 | No |
| 56 | KLHL35 | NA | 10865 | 0.139 | 0.2919 | No | ||
| 57 | IL17A | NA | IL17A Entrez, Source | interleukin 17A | 11336 | 0.116 | 0.2758 | No |
| 58 | CRYGD | NA | CRYGD Entrez, Source | crystallin, gamma D | 11380 | 0.114 | 0.2753 | No |
| 59 | IFNA14 | NA | IFNA14 Entrez, Source | interferon, alpha 14 | 11454 | 0.112 | 0.2737 | No |
| 60 | PRKACG | NA | PRKACG Entrez, Source | protein kinase, cAMP-dependent, catalytic, gamma | 11521 | 0.110 | 0.2723 | No |
| 61 | KCNB2 | NA | KCNB2 Entrez, Source | potassium voltage-gated channel, Shab-related subfamily, member 2 | 11709 | 0.102 | 0.2664 | No |
| 62 | RAG2 | NA | RAG2 Entrez, Source | recombination activating gene 2 | 11872 | 0.095 | 0.2614 | No |
| 63 | THPO | NA | THPO Entrez, Source | thrombopoietin (myeloproliferative leukemia virus oncogene ligand, megakaryocyte growth and development factor) | 12541 | 0.070 | 0.2375 | No |
| 64 | TRGV7 | NA | TRGV7 Entrez, Source | T cell receptor gamma variable 7 | 12575 | 0.068 | 0.2369 | No |
| 65 | LGALS9 | NA | LGALS9 Entrez, Source | lectin, galactoside-binding, soluble, 9 (galectin 9) | 12602 | 0.067 | 0.2366 | No |
| 66 | IQCA1 | NA | 12901 | 0.057 | 0.2262 | No | ||
| 67 | DDX25 | NA | DDX25 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 25 | 13201 | 0.045 | 0.2156 | No |
| 68 | HOXB7 | NA | HOXB7 Entrez, Source | homeobox B7 | 13296 | 0.041 | 0.2126 | No |
| 69 | USP9Y | NA | USP9Y Entrez, Source | ubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) | 13363 | 0.039 | 0.2105 | No |
| 70 | CLEC7A | NA | CLEC7A Entrez, Source | C-type lectin domain family 7, member A | 13483 | 0.034 | 0.2065 | No |
| 71 | INE1 | NA | INE1 Entrez, Source | inactivation escape 1 | 13514 | 0.033 | 0.2057 | No |
| 72 | CD34 | NA | CD34 Entrez, Source | CD34 molecule | 13577 | 0.030 | 0.2037 | No |
| 73 | SPO11 | NA | SPO11 Entrez, Source | SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae) | 13763 | 0.024 | 0.1971 | No |
| 74 | TRPA1 | NA | TRPA1 Entrez, Source | transient receptor potential cation channel, subfamily A, member 1 | 13831 | 0.021 | 0.1949 | No |
| 75 | HAAO | NA | HAAO Entrez, Source | 3-hydroxyanthranilate 3,4-dioxygenase | 13959 | 0.017 | 0.1903 | No |
| 76 | CMKLR1 | NA | CMKLR1 Entrez, Source | chemokine-like receptor 1 | 14027 | 0.014 | 0.1880 | No |
| 77 | ARHGAP15 | NA | ARHGAP15 Entrez, Source | Rho GTPase activating protein 15 | 14093 | 0.012 | 0.1857 | No |
| 78 | UPK1B | NA | UPK1B Entrez, Source | uroplakin 1B | 14216 | 0.008 | 0.1813 | No |
| 79 | CTLA4 | NA | CTLA4 Entrez, Source | cytotoxic T-lymphocyte-associated protein 4 | 14412 | 0.001 | 0.1742 | No |
| 80 | TACR3 | NA | TACR3 Entrez, Source | tachykinin receptor 3 | 14940 | -0.019 | 0.1550 | No |
| 81 | CHRNA6 | NA | CHRNA6 Entrez, Source | cholinergic receptor, nicotinic, alpha 6 | 15092 | -0.025 | 0.1497 | No |
| 82 | CDX1 | NA | CDX1 Entrez, Source | caudal type homeobox transcription factor 1 | 15259 | -0.031 | 0.1439 | No |
| 83 | IL5 | NA | IL5 Entrez, Source | interleukin 5 (colony-stimulating factor, eosinophil) | 15268 | -0.032 | 0.1439 | No |
| 84 | NEUROD4 | NA | NEUROD4 Entrez, Source | neurogenic differentiation 4 | 15357 | -0.035 | 0.1410 | No |
| 85 | SLURP1 | NA | SLURP1 Entrez, Source | secreted LY6/PLAUR domain containing 1 | 15581 | -0.044 | 0.1332 | No |
| 86 | TP53TG5 | NA | 16201 | -0.069 | 0.1111 | No | ||
| 87 | CLDN6 | NA | CLDN6 Entrez, Source | claudin 6 | 16248 | -0.071 | 0.1101 | No |
| 88 | LOC652147 | NA | LOC652147 Entrez, Source | - | 16438 | -0.079 | 0.1039 | No |
| 89 | C7orf63 | NA | 16691 | -0.089 | 0.0955 | No | ||
| 90 | HIST1H3E | NA | HIST1H3E Entrez, Source | histone cluster 1, H3e | 16714 | -0.090 | 0.0955 | No |
| 91 | APOH | NA | APOH Entrez, Source | apolipoprotein H (beta-2-glycoprotein I) | 16916 | -0.098 | 0.0891 | No |
| 92 | ASMT | NA | ASMT Entrez, Source | acetylserotonin O-methyltransferase | 16925 | -0.098 | 0.0897 | No |
| 93 | LRTM1 | NA | LRTM1 Entrez, Source | leucine-rich repeats and transmembrane domains 1 | 17341 | -0.117 | 0.0756 | No |
| 94 | BTG4 | NA | BTG4 Entrez, Source | B-cell translocation gene 4 | 17352 | -0.117 | 0.0764 | No |
| 95 | KCNH7 | NA | KCNH7 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 7 | 17362 | -0.118 | 0.0772 | No |
| 96 | PHOX2B | NA | PHOX2B Entrez, Source | paired-like homeobox 2b | 17549 | -0.126 | 0.0715 | No |
| 97 | DAZL | NA | DAZL Entrez, Source | deleted in azoospermia-like | 17701 | -0.134 | 0.0673 | No |
| 98 | CLDN14 | NA | CLDN14 Entrez, Source | claudin 14 | 17779 | -0.139 | 0.0658 | No |
| 99 | CDKL5 | NA | CDKL5 Entrez, Source | cyclin-dependent kinase-like 5 | 17788 | -0.139 | 0.0668 | No |
| 100 | GJA5 | NA | GJA5 Entrez, Source | gap junction protein, alpha 5, 40kDa (connexin 40) | 18191 | -0.165 | 0.0536 | No |
| 101 | CDH22 | NA | CDH22 Entrez, Source | cadherin-like 22 | 18597 | -0.193 | 0.0406 | No |
| 102 | TRPV6 | NA | TRPV6 Entrez, Source | transient receptor potential cation channel, subfamily V, member 6 | 18921 | -0.220 | 0.0308 | No |
| 103 | SLCO1A2 | NA | SLCO1A2 Entrez, Source | solute carrier organic anion transporter family, member 1A2 | 19188 | -0.243 | 0.0234 | No |
| 104 | PIP | NA | PIP Entrez, Source | prolactin-induced protein | 19201 | -0.244 | 0.0253 | No |
| 105 | GML | NA | GML Entrez, Source | GPI anchored molecule like protein | 19427 | -0.263 | 0.0195 | No |
| 106 | PAEP | NA | PAEP Entrez, Source | progestagen-associated endometrial protein (placental protein 14, pregnancy-associated endometrial alpha-2-globulin, alpha uterine protein) | 19460 | -0.266 | 0.0209 | No |
| 107 | MYBPC1 | NA | MYBPC1 Entrez, Source | myosin binding protein C, slow type | 19575 | -0.278 | 0.0194 | No |
| 108 | RHOH | NA | RHOH Entrez, Source | ras homolog gene family, member H | 19652 | -0.285 | 0.0193 | No |
| 109 | SGCA | NA | SGCA Entrez, Source | sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) | 20289 | -0.354 | -0.0007 | No |
| 110 | TLX3 | NA | TLX3 Entrez, Source | T-cell leukemia homeobox 3 | 20332 | -0.360 | 0.0012 | No |
| 111 | RPP25 | NA | RPP25 Entrez, Source | ribonuclease P 25kDa subunit | 21277 | -0.493 | -0.0287 | No |
| 112 | ZNF780B | NA | ZNF780B Entrez, Source | zinc finger protein 780B | 21359 | -0.508 | -0.0268 | No |
| 113 | FCGBP | NA | FCGBP Entrez, Source | Fc fragment of IgG binding protein | 21504 | -0.536 | -0.0270 | No |
| 114 | DENND1C | NA | DENND1C Entrez, Source | DENN/MADD domain containing 1C | 22056 | -0.639 | -0.0411 | No |
| 115 | CHD5 | NA | CHD5 Entrez, Source | chromodomain helicase DNA binding protein 5 | 22061 | -0.640 | -0.0351 | No |
| 116 | RPS2P45 | NA | 22235 | -0.672 | -0.0350 | No | ||
| 117 | KLK8 | NA | KLK8 Entrez, Source | kallikrein 8 (neuropsin/ovasin) | 22369 | -0.699 | -0.0332 | No |
| 118 | CDH13 | NA | CDH13 Entrez, Source | cadherin 13, H-cadherin (heart) | 23118 | -0.862 | -0.0524 | No |
| 119 | ETNK2 | NA | ETNK2 Entrez, Source | ethanolamine kinase 2 | 23351 | -0.914 | -0.0522 | No |
| 120 | RLN1 | NA | RLN1 Entrez, Source | relaxin 1 | 23673 | -0.999 | -0.0544 | No |
| 121 | SHANK1 | NA | SHANK1 Entrez, Source | SH3 and multiple ankyrin repeat domains 1 | 24100 | -1.118 | -0.0594 | No |
| 122 | OSR2 | NA | OSR2 Entrez, Source | odd-skipped related 2 (Drosophila) | 24565 | -1.268 | -0.0642 | No |
| 123 | SLC16A6 | NA | SLC16A6 Entrez, Source | solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 24660 | -1.296 | -0.0553 | No |
| 124 | SLC43A1 | NA | SLC43A1 Entrez, Source | solute carrier family 43, member 1 | 24844 | -1.358 | -0.0490 | No |
| 125 | HMGA2 | NA | HMGA2 Entrez, Source | high mobility group AT-hook 2 | 25163 | -1.488 | -0.0464 | No |
| 126 | CCL27 | NA | CCL27 Entrez, Source | chemokine (C-C motif) ligand 27 | 25417 | -1.600 | -0.0403 | No |
| 127 | ISYNA1 | NA | ISYNA1 Entrez, Source | - | 25481 | -1.631 | -0.0270 | No |
| 128 | AIM2 | NA | AIM2 Entrez, Source | absent in melanoma 2 | 25899 | -1.855 | -0.0245 | No |
| 129 | MTMR8 | NA | MTMR8 Entrez, Source | myotubularin related protein 8 | 26665 | -2.459 | -0.0290 | No |
| 130 | RCN3 | NA | RCN3 Entrez, Source | reticulocalbin 3, EF-hand calcium binding domain | 26680 | -2.475 | -0.0058 | No |
| 131 | VAV1 | NA | VAV1 Entrez, Source | vav 1 oncogene | 27019 | -3.087 | 0.0114 | No |