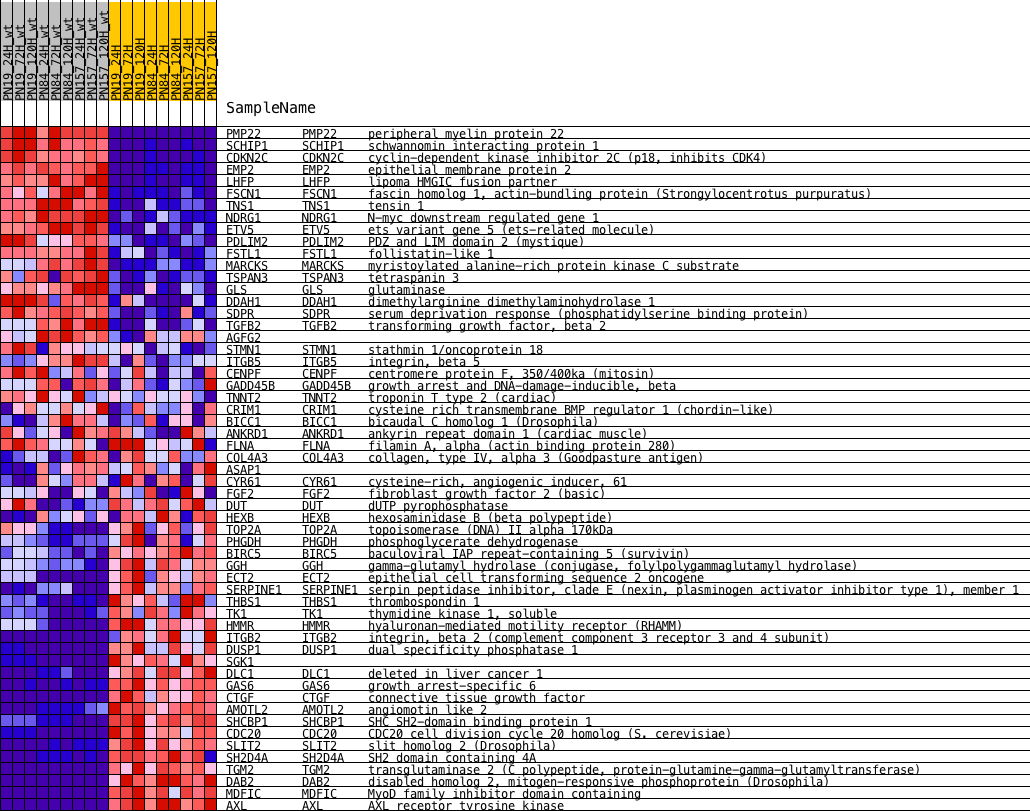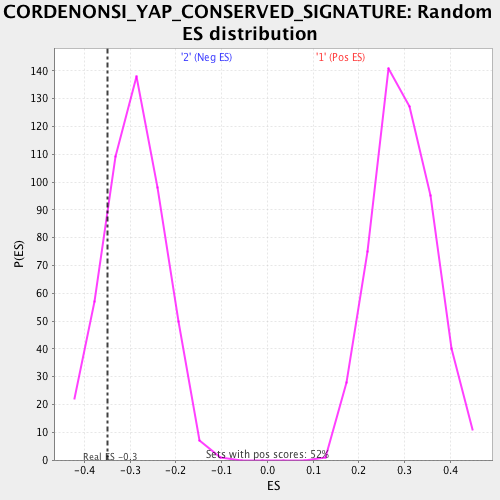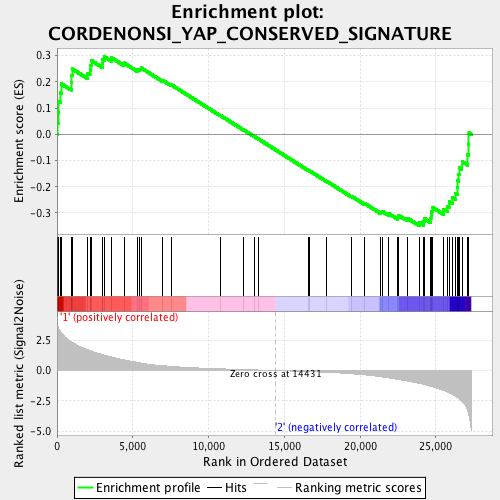
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | CORDENONSI_YAP_CONSERVED_SIGNATURE |
| Enrichment Score (ES) | -0.3497458 |
| Normalized Enrichment Score (NES) | -1.2000318 |
| Nominal p-value | 0.17842324 |
| FDR q-value | 0.23659223 |
| FWER p-Value | 0.918 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PMP22 | NA | PMP22 Entrez, Source | peripheral myelin protein 22 | 41 | 3.713 | 0.0423 | No |
| 2 | SCHIP1 | NA | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 66 | 3.590 | 0.0838 | No |
| 3 | CDKN2C | NA | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 93 | 3.507 | 0.1242 | No |
| 4 | EMP2 | NA | EMP2 Entrez, Source | epithelial membrane protein 2 | 220 | 3.236 | 0.1578 | No |
| 5 | LHFP | NA | LHFP Entrez, Source | lipoma HMGIC fusion partner | 265 | 3.144 | 0.1933 | No |
| 6 | FSCN1 | NA | FSCN1 Entrez, Source | fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | 929 | 2.366 | 0.1969 | No |
| 7 | TNS1 | NA | TNS1 Entrez, Source | tensin 1 | 942 | 2.360 | 0.2243 | No |
| 8 | NDRG1 | NA | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 988 | 2.321 | 0.2501 | No |
| 9 | ETV5 | NA | ETV5 Entrez, Source | ets variant gene 5 (ets-related molecule) | 1991 | 1.731 | 0.2337 | No |
| 10 | PDLIM2 | NA | PDLIM2 Entrez, Source | PDZ and LIM domain 2 (mystique) | 2171 | 1.645 | 0.2466 | No |
| 11 | FSTL1 | NA | FSTL1 Entrez, Source | follistatin-like 1 | 2211 | 1.629 | 0.2644 | No |
| 12 | MARCKS | NA | MARCKS Entrez, Source | myristoylated alanine-rich protein kinase C substrate | 2293 | 1.599 | 0.2803 | No |
| 13 | TSPAN3 | NA | TSPAN3 Entrez, Source | tetraspanin 3 | 2973 | 1.318 | 0.2710 | No |
| 14 | GLS | NA | GLS Entrez, Source | glutaminase | 2991 | 1.313 | 0.2858 | No |
| 15 | DDAH1 | NA | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 3132 | 1.264 | 0.2956 | No |
| 16 | SDPR | NA | SDPR Entrez, Source | serum deprivation response (phosphatidylserine binding protein) | 3594 | 1.120 | 0.2919 | No |
| 17 | TGFB2 | NA | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 4414 | 0.880 | 0.2723 | No |
| 18 | AGFG2 | NA | 5303 | 0.661 | 0.2475 | No | ||
| 19 | STMN1 | NA | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 5464 | 0.626 | 0.2491 | No |
| 20 | ITGB5 | NA | ITGB5 Entrez, Source | integrin, beta 5 | 5536 | 0.610 | 0.2536 | No |
| 21 | CENPF | NA | CENPF Entrez, Source | centromere protein F, 350/400ka (mitosin) | 6984 | 0.399 | 0.2053 | No |
| 22 | GADD45B | NA | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 7552 | 0.340 | 0.1885 | No |
| 23 | TNNT2 | NA | TNNT2 Entrez, Source | troponin T type 2 (cardiac) | 10748 | 0.144 | 0.0731 | No |
| 24 | CRIM1 | NA | CRIM1 Entrez, Source | cysteine rich transmembrane BMP regulator 1 (chordin-like) | 12325 | 0.078 | 0.0162 | No |
| 25 | BICC1 | NA | BICC1 Entrez, Source | bicaudal C homolog 1 (Drosophila) | 13045 | 0.051 | -0.0096 | No |
| 26 | ANKRD1 | NA | ANKRD1 Entrez, Source | ankyrin repeat domain 1 (cardiac muscle) | 13268 | 0.042 | -0.0172 | No |
| 27 | FLNA | NA | FLNA Entrez, Source | filamin A, alpha (actin binding protein 280) | 16587 | -0.084 | -0.1379 | No |
| 28 | COL4A3 | NA | COL4A3 Entrez, Source | collagen, type IV, alpha 3 (Goodpasture antigen) | 16660 | -0.087 | -0.1395 | No |
| 29 | ASAP1 | NA | 17762 | -0.138 | -0.1782 | No | ||
| 30 | CYR61 | NA | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 19423 | -0.263 | -0.2360 | No |
| 31 | FGF2 | NA | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 20293 | -0.354 | -0.2637 | No |
| 32 | DUT | NA | DUT Entrez, Source | dUTP pyrophosphatase | 21369 | -0.509 | -0.2971 | No |
| 33 | HEXB | NA | HEXB Entrez, Source | hexosaminidase B (beta polypeptide) | 21446 | -0.525 | -0.2937 | No |
| 34 | TOP2A | NA | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 21883 | -0.606 | -0.3025 | No |
| 35 | PHGDH | NA | PHGDH Entrez, Source | phosphoglycerate dehydrogenase | 22483 | -0.726 | -0.3159 | No |
| 36 | BIRC5 | NA | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 22533 | -0.737 | -0.3090 | No |
| 37 | GGH | NA | GGH Entrez, Source | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | 23100 | -0.858 | -0.3196 | No |
| 38 | ECT2 | NA | ECT2 Entrez, Source | epithelial cell transforming sequence 2 oncogene | 23923 | -1.071 | -0.3371 | Yes |
| 39 | SERPINE1 | NA | SERPINE1 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 24193 | -1.147 | -0.3334 | Yes |
| 40 | THBS1 | NA | THBS1 Entrez, Source | thrombospondin 1 | 24222 | -1.157 | -0.3208 | Yes |
| 41 | TK1 | NA | TK1 Entrez, Source | thymidine kinase 1, soluble | 24625 | -1.283 | -0.3204 | Yes |
| 42 | HMMR | NA | HMMR Entrez, Source | hyaluronan-mediated motility receptor (RHAMM) | 24674 | -1.301 | -0.3068 | Yes |
| 43 | ITGB2 | NA | ITGB2 Entrez, Source | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 24721 | -1.317 | -0.2930 | Yes |
| 44 | DUSP1 | NA | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 24798 | -1.344 | -0.2799 | Yes |
| 45 | SGK1 | NA | 25489 | -1.638 | -0.2859 | Yes | ||
| 46 | DLC1 | NA | DLC1 Entrez, Source | deleted in liver cancer 1 | 25743 | -1.761 | -0.2743 | Yes |
| 47 | GAS6 | NA | GAS6 Entrez, Source | growth arrest-specific 6 | 25863 | -1.837 | -0.2570 | Yes |
| 48 | CTGF | NA | CTGF Entrez, Source | connective tissue growth factor | 26085 | -1.972 | -0.2419 | Yes |
| 49 | AMOTL2 | NA | AMOTL2 Entrez, Source | angiomotin like 2 | 26277 | -2.110 | -0.2240 | Yes |
| 50 | SHCBP1 | NA | SHCBP1 Entrez, Source | SHC SH2-domain binding protein 1 | 26401 | -2.215 | -0.2023 | Yes |
| 51 | CDC20 | NA | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 26422 | -2.230 | -0.1767 | Yes |
| 52 | SLIT2 | NA | SLIT2 Entrez, Source | slit homolog 2 (Drosophila) | 26489 | -2.288 | -0.1522 | Yes |
| 53 | SH2D4A | NA | SH2D4A Entrez, Source | SH2 domain containing 4A | 26528 | -2.322 | -0.1261 | Yes |
| 54 | TGM2 | NA | TGM2 Entrez, Source | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 26716 | -2.524 | -0.1032 | Yes |
| 55 | DAB2 | NA | DAB2 Entrez, Source | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 27101 | -3.363 | -0.0776 | Yes |
| 56 | MDFIC | NA | MDFIC Entrez, Source | MyoD family inhibitor domain containing | 27169 | -3.607 | -0.0375 | Yes |
| 57 | AXL | NA | AXL Entrez, Source | AXL receptor tyrosine kinase | 27177 | -3.671 | 0.0056 | Yes |