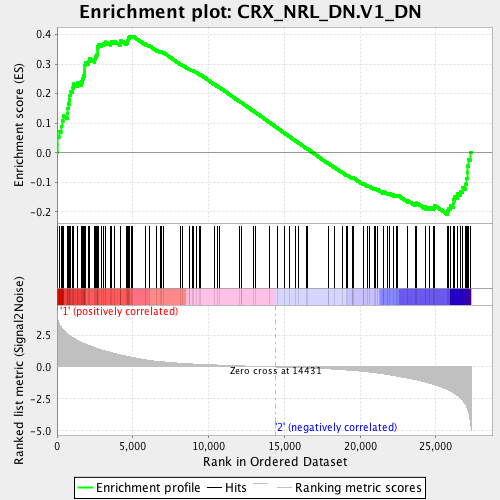
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | CRX_NRL_DN.V1_DN |
| Enrichment Score (ES) | 0.39447355 |
| Normalized Enrichment Score (NES) | 1.461463 |
| Nominal p-value | 0.008032128 |
| FDR q-value | 0.04582918 |
| FWER p-Value | 0.359 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | INHBB | NA | INHBB Entrez, Source | inhibin, beta B (activin AB beta polypeptide) | 5 | 4.070 | 0.0274 | Yes |
| 2 | HES5 | NA | HES5 Entrez, Source | hairy and enhancer of split 5 (Drosophila) | 7 | 4.026 | 0.0546 | Yes |
| 3 | IL17D | NA | IL17D Entrez, Source | interleukin 17D | 127 | 3.407 | 0.0733 | Yes |
| 4 | TNC | NA | TNC Entrez, Source | tenascin C (hexabrachion) | 258 | 3.160 | 0.0900 | Yes |
| 5 | FABP7 | NA | FABP7 Entrez, Source | fatty acid binding protein 7, brain | 323 | 3.044 | 0.1082 | Yes |
| 6 | FRMD6 | NA | FRMD6 Entrez, Source | FERM domain containing 6 | 392 | 2.939 | 0.1256 | Yes |
| 7 | GTF2I | NA | GTF2I Entrez, Source | general transcription factor II, i | 689 | 2.587 | 0.1323 | Yes |
| 8 | JAM3 | NA | JAM3 Entrez, Source | junctional adhesion molecule 3 | 696 | 2.580 | 0.1495 | Yes |
| 9 | TNPO2 | NA | TNPO2 Entrez, Source | transportin 2 (importin 3, karyopherin beta 2b) | 729 | 2.544 | 0.1656 | Yes |
| 10 | PAM | NA | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 791 | 2.484 | 0.1802 | Yes |
| 11 | LRRC4 | NA | LRRC4 Entrez, Source | leucine rich repeat containing 4 | 840 | 2.441 | 0.1950 | Yes |
| 12 | RTKN2 | NA | 907 | 2.386 | 0.2087 | Yes | ||
| 13 | CITED1 | NA | CITED1 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 | 1002 | 2.316 | 0.2209 | Yes |
| 14 | KIF5C | NA | KIF5C Entrez, Source | kinesin family member 5C | 1092 | 2.248 | 0.2329 | Yes |
| 15 | METRN | NA | METRN Entrez, Source | meteorin, glial cell differentiation regulator | 1370 | 2.059 | 0.2367 | Yes |
| 16 | C14orf132 | NA | C14ORF132 Entrez, Source | chromosome 14 open reading frame 132 | 1582 | 1.932 | 0.2420 | Yes |
| 17 | BAMBI | NA | BAMBI Entrez, Source | BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | 1692 | 1.877 | 0.2507 | Yes |
| 18 | TMEM87A | NA | TMEM87A Entrez, Source | transmembrane protein 87A | 1734 | 1.854 | 0.2617 | Yes |
| 19 | BAX | NA | BAX Entrez, Source | BCL2-associated X protein | 1823 | 1.810 | 0.2708 | Yes |
| 20 | PTPN13 | NA | PTPN13 Entrez, Source | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | 1829 | 1.809 | 0.2828 | Yes |
| 21 | SLC26A4 | NA | SLC26A4 Entrez, Source | solute carrier family 26, member 4 | 1834 | 1.807 | 0.2949 | Yes |
| 22 | GGT7 | NA | 1866 | 1.795 | 0.3060 | Yes | ||
| 23 | U2AF1 | NA | U2AF1 Entrez, Source | U2 small nuclear RNA auxiliary factor 1 | 2078 | 1.686 | 0.3096 | Yes |
| 24 | PVRL3 | NA | PVRL3 Entrez, Source | poliovirus receptor-related 3 | 2134 | 1.663 | 0.3189 | Yes |
| 25 | HRASLS | NA | HRASLS Entrez, Source | HRAS-like suppressor | 2473 | 1.516 | 0.3167 | Yes |
| 26 | ENC1 | NA | ENC1 Entrez, Source | ectodermal-neural cortex (with BTB-like domain) | 2528 | 1.494 | 0.3248 | Yes |
| 27 | ERMAP | NA | ERMAP Entrez, Source | erythroblast membrane-associated protein (Scianna blood group) | 2623 | 1.451 | 0.3312 | Yes |
| 28 | TMEM106B | NA | TMEM106B Entrez, Source | transmembrane protein 106B | 2644 | 1.446 | 0.3403 | Yes |
| 29 | CCDC23 | NA | CCDC23 Entrez, Source | coiled-coil domain containing 23 | 2646 | 1.446 | 0.3500 | Yes |
| 30 | PYGM | NA | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 2652 | 1.443 | 0.3596 | Yes |
| 31 | ELOVL2 | NA | ELOVL2 Entrez, Source | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 | 2759 | 1.396 | 0.3652 | Yes |
| 32 | CAMK2B | NA | CAMK2B Entrez, Source | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta | 2960 | 1.324 | 0.3668 | Yes |
| 33 | GPR124 | NA | GPR124 Entrez, Source | G protein-coupled receptor 124 | 3080 | 1.284 | 0.3711 | Yes |
| 34 | INPPL1 | NA | INPPL1 Entrez, Source | inositol polyphosphate phosphatase-like 1 | 3212 | 1.236 | 0.3747 | Yes |
| 35 | ECE1 | NA | ECE1 Entrez, Source | endothelin converting enzyme 1 | 3542 | 1.140 | 0.3703 | Yes |
| 36 | FAM134C | NA | 3571 | 1.130 | 0.3769 | Yes | ||
| 37 | GREB1 | NA | GREB1 Entrez, Source | - | 3762 | 1.065 | 0.3772 | Yes |
| 38 | FMN1 | NA | FMN1 Entrez, Source | formin 1 | 4156 | 0.942 | 0.3691 | Yes |
| 39 | DFNA5 | NA | DFNA5 Entrez, Source | deafness, autosomal dominant 5 | 4198 | 0.932 | 0.3739 | Yes |
| 40 | XK | NA | XK Entrez, Source | X-linked Kx blood group (McLeod syndrome) | 4199 | 0.932 | 0.3802 | Yes |
| 41 | DNAJC9 | NA | DNAJC9 Entrez, Source | DnaJ (Hsp40) homolog, subfamily C, member 9 | 4606 | 0.841 | 0.3710 | Yes |
| 42 | MFNG | NA | MFNG Entrez, Source | manic fringe homolog (Drosophila) | 4627 | 0.832 | 0.3759 | Yes |
| 43 | SLC25A13 | NA | SLC25A13 Entrez, Source | solute carrier family 25, member 13 (citrin) | 4657 | 0.824 | 0.3804 | Yes |
| 44 | TNNC2 | NA | TNNC2 Entrez, Source | troponin C type 2 (fast) | 4681 | 0.818 | 0.3851 | Yes |
| 45 | SALL3 | NA | SALL3 Entrez, Source | sal-like 3 (Drosophila) | 4716 | 0.806 | 0.3893 | Yes |
| 46 | NTN4 | NA | NTN4 Entrez, Source | netrin 4 | 4796 | 0.790 | 0.3918 | Yes |
| 47 | NIN | NA | NIN Entrez, Source | ninein (GSK3B interacting protein) | 4895 | 0.768 | 0.3934 | Yes |
| 48 | FZD5 | NA | FZD5 Entrez, Source | frizzled homolog 5 (Drosophila) | 5003 | 0.742 | 0.3945 | Yes |
| 49 | NTNG2 | NA | NTNG2 Entrez, Source | netrin G2 | 5823 | 0.557 | 0.3681 | No |
| 50 | CDH20 | NA | CDH20 Entrez, Source | cadherin 20, type 2 | 6088 | 0.513 | 0.3619 | No |
| 51 | RNF25 | NA | RNF25 Entrez, Source | ring finger protein 25 | 6574 | 0.444 | 0.3471 | No |
| 52 | PRMT1 | NA | PRMT1 Entrez, Source | protein arginine methyltransferase 1 | 6833 | 0.415 | 0.3404 | No |
| 53 | D4S234E | NA | D4S234E Entrez, Source | - | 6856 | 0.412 | 0.3424 | No |
| 54 | MYOCD | NA | MYOCD Entrez, Source | myocardin | 7014 | 0.396 | 0.3393 | No |
| 55 | CHRNB4 | NA | CHRNB4 Entrez, Source | cholinergic receptor, nicotinic, beta 4 | 8139 | 0.289 | 0.3000 | No |
| 56 | RBP4 | NA | RBP4 Entrez, Source | retinol binding protein 4, plasma | 8284 | 0.278 | 0.2966 | No |
| 57 | C11orf52 | NA | C11ORF52 Entrez, Source | chromosome 11 open reading frame 52 | 8718 | 0.247 | 0.2823 | No |
| 58 | ABCC9 | NA | ABCC9 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 | 8766 | 0.243 | 0.2822 | No |
| 59 | SMPDL3A | NA | SMPDL3A Entrez, Source | sphingomyelin phosphodiesterase, acid-like 3A | 8919 | 0.234 | 0.2782 | No |
| 60 | ARPC3 | NA | ARPC3 Entrez, Source | actin related protein 2/3 complex, subunit 3, 21kDa | 8966 | 0.232 | 0.2781 | No |
| 61 | CHRNA3 | NA | CHRNA3 Entrez, Source | cholinergic receptor, nicotinic, alpha 3 | 9167 | 0.221 | 0.2723 | No |
| 62 | CCKBR | NA | CCKBR Entrez, Source | cholecystokinin B receptor | 9421 | 0.207 | 0.2644 | No |
| 63 | MYBPH | NA | MYBPH Entrez, Source | myosin binding protein H | 9432 | 0.207 | 0.2654 | No |
| 64 | CNGB3 | NA | CNGB3 Entrez, Source | cyclic nucleotide gated channel beta 3 | 10413 | 0.160 | 0.2305 | No |
| 65 | BSPRY | NA | BSPRY Entrez, Source | B-box and SPRY domain containing | 10565 | 0.153 | 0.2259 | No |
| 66 | CPLX2 | NA | CPLX2 Entrez, Source | complexin 2 | 10726 | 0.145 | 0.2210 | No |
| 67 | GABRA2 | NA | GABRA2 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 2 | 12036 | 0.089 | 0.1735 | No |
| 68 | EYA1 | NA | EYA1 Entrez, Source | eyes absent homolog 1 (Drosophila) | 12135 | 0.085 | 0.1705 | No |
| 69 | MTNR1A | NA | MTNR1A Entrez, Source | melatonin receptor 1A | 12152 | 0.085 | 0.1705 | No |
| 70 | SPINT1 | NA | SPINT1 Entrez, Source | serine peptidase inhibitor, Kunitz type 1 | 12941 | 0.055 | 0.1419 | No |
| 71 | NMBR | NA | NMBR Entrez, Source | neuromedin B receptor | 13117 | 0.048 | 0.1358 | No |
| 72 | RASGRF2 | NA | RASGRF2 Entrez, Source | Ras protein-specific guanine nucleotide-releasing factor 2 | 14023 | 0.014 | 0.1026 | No |
| 73 | DPRX | NA | DPRX Entrez, Source | divergent-paired related homeobox | 14549 | -0.004 | 0.0834 | No |
| 74 | TSPAN33 | NA | TSPAN33 Entrez, Source | tetraspanin 33 | 15029 | -0.022 | 0.0659 | No |
| 75 | KLHL1 | NA | KLHL1 Entrez, Source | kelch-like 1 (Drosophila) | 15343 | -0.035 | 0.0546 | No |
| 76 | GRIK2 | NA | GRIK2 Entrez, Source | glutamate receptor, ionotropic, kainate 2 | 15758 | -0.051 | 0.0398 | No |
| 77 | AP2B1 | NA | AP2B1 Entrez, Source | adaptor-related protein complex 2, beta 1 subunit | 15912 | -0.056 | 0.0345 | No |
| 78 | PDIA3 | NA | PDIA3 Entrez, Source | protein disulfide isomerase family A, member 3 | 16486 | -0.080 | 0.0140 | No |
| 79 | HS3ST1 | NA | HS3ST1 Entrez, Source | heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | 16530 | -0.082 | 0.0130 | No |
| 80 | GDPD3 | NA | GDPD3 Entrez, Source | glycerophosphodiester phosphodiesterase domain containing 3 | 17880 | -0.146 | -0.0356 | No |
| 81 | RREB1 | NA | RREB1 Entrez, Source | ras responsive element binding protein 1 | 17905 | -0.148 | -0.0355 | No |
| 82 | DDX3Y | NA | DDX3Y Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 18301 | -0.172 | -0.0489 | No |
| 83 | ADAT2 | NA | 18819 | -0.211 | -0.0664 | No | ||
| 84 | ZNF316 | NA | ZNF316 Entrez, Source | zinc finger protein 316 | 19119 | -0.237 | -0.0758 | No |
| 85 | TNFRSF12A | NA | TNFRSF12A Entrez, Source | tumor necrosis factor receptor superfamily, member 12A | 19170 | -0.241 | -0.0760 | No |
| 86 | LSM12 | NA | LSM12 Entrez, Source | LSM12 homolog (S. cerevisiae) | 19487 | -0.268 | -0.0858 | No |
| 87 | ZNF146 | NA | ZNF146 Entrez, Source | zinc finger protein 146 | 19505 | -0.270 | -0.0846 | No |
| 88 | CLIC4 | NA | CLIC4 Entrez, Source | chloride intracellular channel 4 | 19546 | -0.275 | -0.0842 | No |
| 89 | CAP1 | NA | CAP1 Entrez, Source | CAP, adenylate cyclase-associated protein 1 (yeast) | 20201 | -0.341 | -0.1059 | No |
| 90 | PRDM1 | NA | PRDM1 Entrez, Source | PR domain containing 1, with ZNF domain | 20217 | -0.343 | -0.1042 | No |
| 91 | CRYBG3 | NA | 20471 | -0.377 | -0.1109 | No | ||
| 92 | NR4A1 | NA | NR4A1 Entrez, Source | nuclear receptor subfamily 4, group A, member 1 | 20585 | -0.391 | -0.1124 | No |
| 93 | MFSD4 | NA | MFSD4 Entrez, Source | major facilitator superfamily domain containing 4 | 20937 | -0.443 | -0.1223 | No |
| 94 | RXRG | NA | RXRG Entrez, Source | retinoid X receptor, gamma | 20994 | -0.451 | -0.1213 | No |
| 95 | ATOH7 | NA | ATOH7 Entrez, Source | atonal homolog 7 (Drosophila) | 21128 | -0.472 | -0.1230 | No |
| 96 | RASA2 | NA | RASA2 Entrez, Source | RAS p21 protein activator 2 | 21520 | -0.538 | -0.1337 | No |
| 97 | ACBD6 | NA | ACBD6 Entrez, Source | acyl-Coenzyme A binding domain containing 6 | 21534 | -0.540 | -0.1306 | No |
| 98 | HEBP1 | NA | HEBP1 Entrez, Source | heme binding protein 1 | 21816 | -0.595 | -0.1369 | No |
| 99 | NFKB1 | NA | NFKB1 Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 21956 | -0.621 | -0.1378 | No |
| 100 | SGCE | NA | SGCE Entrez, Source | sarcoglycan, epsilon | 22189 | -0.663 | -0.1418 | No |
| 101 | INSM2 | NA | INSM2 Entrez, Source | insulinoma-associated 2 | 22377 | -0.702 | -0.1439 | No |
| 102 | C19orf39 | NA | C19ORF39 Entrez, Source | chromosome 19 open reading frame 39 | 22475 | -0.723 | -0.1426 | No |
| 103 | CTBS | NA | CTBS Entrez, Source | chitobiase, di-N-acetyl- | 23125 | -0.863 | -0.1606 | No |
| 104 | PTER | NA | PTER Entrez, Source | phosphotriesterase related | 23633 | -0.988 | -0.1725 | No |
| 105 | A2LD1 | NA | 23736 | -1.016 | -0.1694 | No | ||
| 106 | TRAPPC3 | NA | TRAPPC3 Entrez, Source | trafficking protein particle complex 3 | 24297 | -1.178 | -0.1820 | No |
| 107 | RGL3 | NA | RGL3 Entrez, Source | ral guanine nucleotide dissociation stimulator-like 3 | 24562 | -1.267 | -0.1831 | No |
| 108 | GABRA3 | NA | GABRA3 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | 24833 | -1.355 | -0.1839 | No |
| 109 | ASPH | NA | ASPH Entrez, Source | aspartate beta-hydroxylase | 24917 | -1.385 | -0.1775 | No |
| 110 | ELOVL7 | NA | ELOVL7 Entrez, Source | ELOVL family member 7, elongation of long chain fatty acids (yeast) | 25758 | -1.771 | -0.1964 | No |
| 111 | CLPB | NA | CLPB Entrez, Source | ClpB caseinolytic peptidase B homolog (E. coli) | 25852 | -1.827 | -0.1875 | No |
| 112 | MCART1 | NA | MCART1 Entrez, Source | mitochondrial carrier triple repeat 1 | 25959 | -1.890 | -0.1786 | No |
| 113 | RAB27B | NA | RAB27B Entrez, Source | RAB27B, member RAS oncogene family | 26165 | -2.039 | -0.1723 | No |
| 114 | QSOX1 | NA | 26171 | -2.043 | -0.1586 | No | ||
| 115 | RNF149 | NA | RNF149 Entrez, Source | ring finger protein 149 | 26232 | -2.076 | -0.1468 | No |
| 116 | ZNF600 | NA | ZNF600 Entrez, Source | zinc finger protein 600 | 26405 | -2.217 | -0.1381 | No |
| 117 | TMEM158 | NA | TMEM158 Entrez, Source | transmembrane protein 158 | 26610 | -2.396 | -0.1293 | No |
| 118 | SDC4 | NA | SDC4 Entrez, Source | syndecan 4 (amphiglycan, ryudocan) | 26750 | -2.560 | -0.1171 | No |
| 119 | WWC1 | NA | WWC1 Entrez, Source | WW, C2 and coiled-coil domain containing 1 | 26975 | -2.960 | -0.1053 | No |
| 120 | MINA | NA | MINA Entrez, Source | MYC induced nuclear antigen | 27005 | -3.051 | -0.0857 | No |
| 121 | RERG | NA | RERG Entrez, Source | RAS-like, estrogen-regulated, growth inhibitor | 27069 | -3.233 | -0.0661 | No |
| 122 | RBP7 | NA | RBP7 Entrez, Source | retinol binding protein 7, cellular | 27090 | -3.321 | -0.0443 | No |
| 123 | ADRB2 | NA | ADRB2 Entrez, Source | adrenergic, beta-2-, receptor, surface | 27138 | -3.500 | -0.0223 | No |
| 124 | EGR1 | NA | EGR1 Entrez, Source | early growth response 1 | 27274 | -4.330 | 0.0020 | No |

