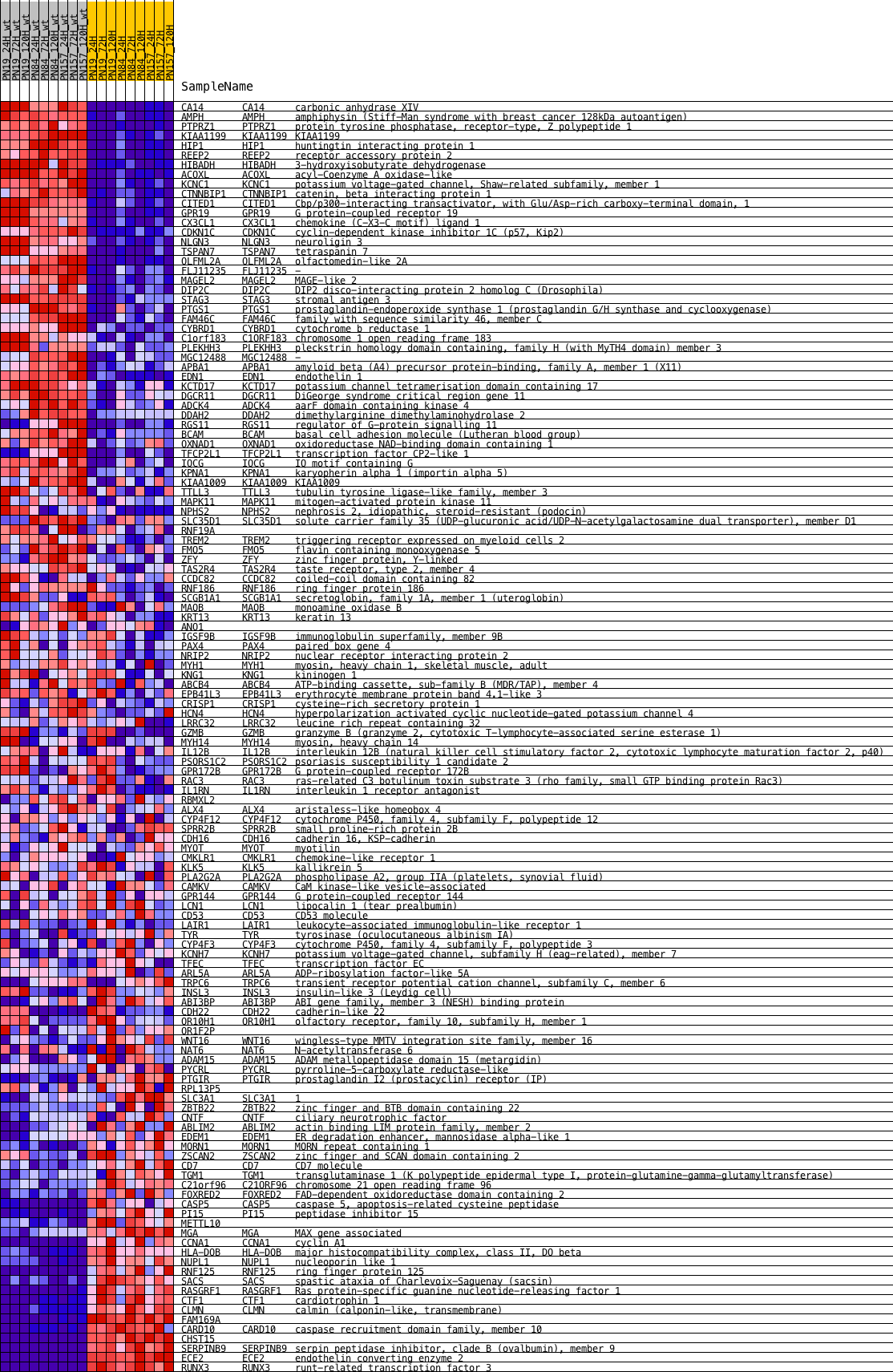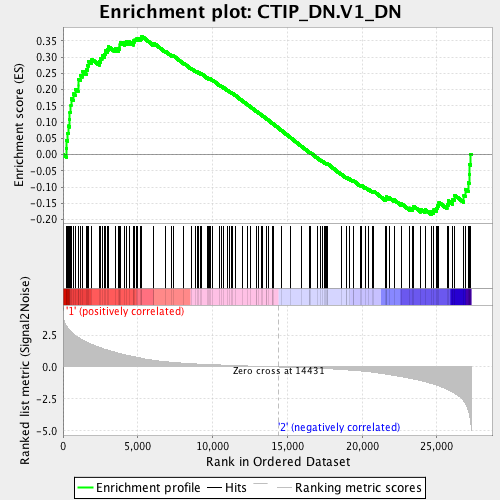
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | CTIP_DN.V1_DN |
| Enrichment Score (ES) | 0.3648695 |
| Normalized Enrichment Score (NES) | 1.4731383 |
| Nominal p-value | 0.00591716 |
| FDR q-value | 0.042541757 |
| FWER p-Value | 0.327 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CA14 | NA | CA14 Entrez, Source | carbonic anhydrase XIV | 205 | 3.262 | 0.0187 | Yes |
| 2 | AMPH | NA | AMPH Entrez, Source | amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen) | 217 | 3.243 | 0.0443 | Yes |
| 3 | PTPRZ1 | NA | PTPRZ1 Entrez, Source | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | 294 | 3.095 | 0.0664 | Yes |
| 4 | KIAA1199 | NA | KIAA1199 Entrez, Source | KIAA1199 | 353 | 2.993 | 0.0884 | Yes |
| 5 | HIP1 | NA | HIP1 Entrez, Source | huntingtin interacting protein 1 | 445 | 2.875 | 0.1081 | Yes |
| 6 | REEP2 | NA | REEP2 Entrez, Source | receptor accessory protein 2 | 455 | 2.863 | 0.1308 | Yes |
| 7 | HIBADH | NA | HIBADH Entrez, Source | 3-hydroxyisobutyrate dehydrogenase | 523 | 2.757 | 0.1505 | Yes |
| 8 | ACOXL | NA | ACOXL Entrez, Source | acyl-Coenzyme A oxidase-like | 531 | 2.743 | 0.1723 | Yes |
| 9 | KCNC1 | NA | KCNC1 Entrez, Source | potassium voltage-gated channel, Shaw-related subfamily, member 1 | 698 | 2.579 | 0.1869 | Yes |
| 10 | CTNNBIP1 | NA | CTNNBIP1 Entrez, Source | catenin, beta interacting protein 1 | 845 | 2.437 | 0.2011 | Yes |
| 11 | CITED1 | NA | CITED1 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 | 1002 | 2.316 | 0.2140 | Yes |
| 12 | GPR19 | NA | GPR19 Entrez, Source | G protein-coupled receptor 19 | 1003 | 2.316 | 0.2326 | Yes |
| 13 | CX3CL1 | NA | CX3CL1 Entrez, Source | chemokine (C-X3-C motif) ligand 1 | 1151 | 2.210 | 0.2450 | Yes |
| 14 | CDKN1C | NA | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 1317 | 2.089 | 0.2557 | Yes |
| 15 | NLGN3 | NA | NLGN3 Entrez, Source | neuroligin 3 | 1570 | 1.940 | 0.2621 | Yes |
| 16 | TSPAN7 | NA | TSPAN7 Entrez, Source | tetraspanin 7 | 1625 | 1.909 | 0.2754 | Yes |
| 17 | OLFML2A | NA | OLFML2A Entrez, Source | olfactomedin-like 2A | 1700 | 1.871 | 0.2877 | Yes |
| 18 | FLJ11235 | NA | FLJ11235 Entrez, Source | - | 1925 | 1.764 | 0.2937 | Yes |
| 19 | MAGEL2 | NA | MAGEL2 Entrez, Source | MAGE-like 2 | 2414 | 1.540 | 0.2881 | Yes |
| 20 | DIP2C | NA | DIP2C Entrez, Source | DIP2 disco-interacting protein 2 homolog C (Drosophila) | 2515 | 1.500 | 0.2965 | Yes |
| 21 | STAG3 | NA | STAG3 Entrez, Source | stromal antigen 3 | 2603 | 1.458 | 0.3050 | Yes |
| 22 | PTGS1 | NA | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 2791 | 1.388 | 0.3093 | Yes |
| 23 | FAM46C | NA | FAM46C Entrez, Source | family with sequence similarity 46, member C | 2818 | 1.380 | 0.3194 | Yes |
| 24 | CYBRD1 | NA | CYBRD1 Entrez, Source | cytochrome b reductase 1 | 2990 | 1.313 | 0.3237 | Yes |
| 25 | C1orf183 | NA | C1ORF183 Entrez, Source | chromosome 1 open reading frame 183 | 3040 | 1.296 | 0.3323 | Yes |
| 26 | PLEKHH3 | NA | PLEKHH3 Entrez, Source | pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 | 3478 | 1.161 | 0.3256 | Yes |
| 27 | MGC12488 | NA | MGC12488 Entrez, Source | - | 3705 | 1.081 | 0.3260 | Yes |
| 28 | APBA1 | NA | APBA1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family A, member 1 (X11) | 3801 | 1.051 | 0.3309 | Yes |
| 29 | EDN1 | NA | EDN1 Entrez, Source | endothelin 1 | 3804 | 1.049 | 0.3393 | Yes |
| 30 | KCTD17 | NA | KCTD17 Entrez, Source | potassium channel tetramerisation domain containing 17 | 3844 | 1.037 | 0.3462 | Yes |
| 31 | DGCR11 | NA | DGCR11 Entrez, Source | DiGeorge syndrome critical region gene 11 | 4097 | 0.960 | 0.3446 | Yes |
| 32 | ADCK4 | NA | ADCK4 Entrez, Source | aarF domain containing kinase 4 | 4224 | 0.926 | 0.3474 | Yes |
| 33 | DDAH2 | NA | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 4426 | 0.877 | 0.3471 | Yes |
| 34 | RGS11 | NA | RGS11 Entrez, Source | regulator of G-protein signalling 11 | 4691 | 0.814 | 0.3439 | Yes |
| 35 | BCAM | NA | BCAM Entrez, Source | basal cell adhesion molecule (Lutheran blood group) | 4702 | 0.810 | 0.3501 | Yes |
| 36 | OXNAD1 | NA | OXNAD1 Entrez, Source | oxidoreductase NAD-binding domain containing 1 | 4798 | 0.790 | 0.3529 | Yes |
| 37 | TFCP2L1 | NA | TFCP2L1 Entrez, Source | transcription factor CP2-like 1 | 4880 | 0.771 | 0.3562 | Yes |
| 38 | IQCG | NA | IQCG Entrez, Source | IQ motif containing G | 5007 | 0.741 | 0.3575 | Yes |
| 39 | KPNA1 | NA | KPNA1 Entrez, Source | karyopherin alpha 1 (importin alpha 5) | 5158 | 0.705 | 0.3576 | Yes |
| 40 | KIAA1009 | NA | KIAA1009 Entrez, Source | KIAA1009 | 5247 | 0.678 | 0.3599 | Yes |
| 41 | TTLL3 | NA | TTLL3 Entrez, Source | tubulin tyrosine ligase-like family, member 3 | 5259 | 0.674 | 0.3649 | Yes |
| 42 | MAPK11 | NA | MAPK11 Entrez, Source | mitogen-activated protein kinase 11 | 6042 | 0.520 | 0.3403 | No |
| 43 | NPHS2 | NA | NPHS2 Entrez, Source | nephrosis 2, idiopathic, steroid-resistant (podocin) | 6079 | 0.515 | 0.3431 | No |
| 44 | SLC35D1 | NA | SLC35D1 Entrez, Source | solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 | 6867 | 0.411 | 0.3175 | No |
| 45 | RNF19A | NA | 7284 | 0.367 | 0.3051 | No | ||
| 46 | TREM2 | NA | TREM2 Entrez, Source | triggering receptor expressed on myeloid cells 2 | 7364 | 0.358 | 0.3051 | No |
| 47 | FMO5 | NA | FMO5 Entrez, Source | flavin containing monooxygenase 5 | 8075 | 0.294 | 0.2814 | No |
| 48 | ZFY | NA | ZFY Entrez, Source | zinc finger protein, Y-linked | 8605 | 0.256 | 0.2640 | No |
| 49 | TAS2R4 | NA | TAS2R4 Entrez, Source | taste receptor, type 2, member 4 | 8862 | 0.238 | 0.2565 | No |
| 50 | CCDC82 | NA | CCDC82 Entrez, Source | coiled-coil domain containing 82 | 8957 | 0.232 | 0.2549 | No |
| 51 | RNF186 | NA | RNF186 Entrez, Source | ring finger protein 186 | 9066 | 0.226 | 0.2527 | No |
| 52 | SCGB1A1 | NA | SCGB1A1 Entrez, Source | secretoglobin, family 1A, member 1 (uteroglobin) | 9216 | 0.218 | 0.2490 | No |
| 53 | MAOB | NA | MAOB Entrez, Source | monoamine oxidase B | 9285 | 0.215 | 0.2482 | No |
| 54 | KRT13 | NA | KRT13 Entrez, Source | keratin 13 | 9639 | 0.195 | 0.2368 | No |
| 55 | ANO1 | NA | 9757 | 0.190 | 0.2341 | No | ||
| 56 | IGSF9B | NA | IGSF9B Entrez, Source | immunoglobulin superfamily, member 9B | 9806 | 0.188 | 0.2338 | No |
| 57 | PAX4 | NA | PAX4 Entrez, Source | paired box gene 4 | 9836 | 0.186 | 0.2342 | No |
| 58 | NRIP2 | NA | NRIP2 Entrez, Source | nuclear receptor interacting protein 2 | 10013 | 0.178 | 0.2292 | No |
| 59 | MYH1 | NA | MYH1 Entrez, Source | myosin, heavy chain 1, skeletal muscle, adult | 10487 | 0.156 | 0.2131 | No |
| 60 | KNG1 | NA | KNG1 Entrez, Source | kininogen 1 | 10587 | 0.152 | 0.2106 | No |
| 61 | ABCB4 | NA | ABCB4 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 4 | 10750 | 0.144 | 0.2058 | No |
| 62 | EPB41L3 | NA | EPB41L3 Entrez, Source | erythrocyte membrane protein band 4.1-like 3 | 10988 | 0.133 | 0.1982 | No |
| 63 | CRISP1 | NA | CRISP1 Entrez, Source | cysteine-rich secretory protein 1 | 11103 | 0.127 | 0.1950 | No |
| 64 | HCN4 | NA | HCN4 Entrez, Source | hyperpolarization activated cyclic nucleotide-gated potassium channel 4 | 11285 | 0.118 | 0.1893 | No |
| 65 | LRRC32 | NA | LRRC32 Entrez, Source | leucine rich repeat containing 32 | 11303 | 0.118 | 0.1896 | No |
| 66 | GZMB | NA | GZMB Entrez, Source | granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | 11501 | 0.110 | 0.1833 | No |
| 67 | MYH14 | NA | MYH14 Entrez, Source | myosin, heavy chain 14 | 11972 | 0.091 | 0.1667 | No |
| 68 | IL12B | NA | IL12B Entrez, Source | interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) | 12329 | 0.078 | 0.1543 | No |
| 69 | PSORS1C2 | NA | PSORS1C2 Entrez, Source | psoriasis susceptibility 1 candidate 2 | 12533 | 0.070 | 0.1474 | No |
| 70 | GPR172B | NA | GPR172B Entrez, Source | G protein-coupled receptor 172B | 12919 | 0.056 | 0.1337 | No |
| 71 | RAC3 | NA | RAC3 Entrez, Source | ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | 12953 | 0.055 | 0.1329 | No |
| 72 | IL1RN | NA | IL1RN Entrez, Source | interleukin 1 receptor antagonist | 13063 | 0.050 | 0.1293 | No |
| 73 | RBMXL2 | NA | 13279 | 0.042 | 0.1217 | No | ||
| 74 | ALX4 | NA | ALX4 Entrez, Source | aristaless-like homeobox 4 | 13362 | 0.039 | 0.1190 | No |
| 75 | CYP4F12 | NA | CYP4F12 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 12 | 13616 | 0.029 | 0.1100 | No |
| 76 | SPRR2B | NA | SPRR2B Entrez, Source | small proline-rich protein 2B | 13619 | 0.029 | 0.1101 | No |
| 77 | CDH16 | NA | CDH16 Entrez, Source | cadherin 16, KSP-cadherin | 13744 | 0.024 | 0.1057 | No |
| 78 | MYOT | NA | MYOT Entrez, Source | myotilin | 13995 | 0.015 | 0.0967 | No |
| 79 | CMKLR1 | NA | CMKLR1 Entrez, Source | chemokine-like receptor 1 | 14027 | 0.014 | 0.0957 | No |
| 80 | KLK5 | NA | KLK5 Entrez, Source | kallikrein 5 | 14099 | 0.012 | 0.0931 | No |
| 81 | PLA2G2A | NA | PLA2G2A Entrez, Source | phospholipase A2, group IIA (platelets, synovial fluid) | 14628 | -0.007 | 0.0738 | No |
| 82 | CAMKV | NA | CAMKV Entrez, Source | CaM kinase-like vesicle-associated | 15227 | -0.030 | 0.0520 | No |
| 83 | GPR144 | NA | GPR144 Entrez, Source | G protein-coupled receptor 144 | 15945 | -0.057 | 0.0261 | No |
| 84 | LCN1 | NA | LCN1 Entrez, Source | lipocalin 1 (tear prealbumin) | 16491 | -0.081 | 0.0067 | No |
| 85 | CD53 | NA | CD53 Entrez, Source | CD53 molecule | 16512 | -0.081 | 0.0067 | No |
| 86 | LAIR1 | NA | LAIR1 Entrez, Source | leukocyte-associated immunoglobulin-like receptor 1 | 16521 | -0.082 | 0.0070 | No |
| 87 | TYR | NA | TYR Entrez, Source | tyrosinase (oculocutaneous albinism IA) | 17010 | -0.102 | -0.0101 | No |
| 88 | CYP4F3 | NA | CYP4F3 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 3 | 17205 | -0.111 | -0.0163 | No |
| 89 | KCNH7 | NA | KCNH7 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 7 | 17362 | -0.118 | -0.0211 | No |
| 90 | TFEC | NA | TFEC Entrez, Source | transcription factor EC | 17495 | -0.124 | -0.0250 | No |
| 91 | ARL5A | NA | ARL5A Entrez, Source | ADP-ribosylation factor-like 5A | 17583 | -0.128 | -0.0272 | No |
| 92 | TRPC6 | NA | TRPC6 Entrez, Source | transient receptor potential cation channel, subfamily C, member 6 | 17634 | -0.131 | -0.0280 | No |
| 93 | INSL3 | NA | INSL3 Entrez, Source | insulin-like 3 (Leydig cell) | 17662 | -0.132 | -0.0279 | No |
| 94 | ABI3BP | NA | ABI3BP Entrez, Source | ABI gene family, member 3 (NESH) binding protein | 17681 | -0.133 | -0.0275 | No |
| 95 | CDH22 | NA | CDH22 Entrez, Source | cadherin-like 22 | 18597 | -0.193 | -0.0596 | No |
| 96 | OR10H1 | NA | OR10H1 Entrez, Source | olfactory receptor, family 10, subfamily H, member 1 | 18978 | -0.225 | -0.0717 | No |
| 97 | OR1F2P | NA | 18987 | -0.226 | -0.0702 | No | ||
| 98 | WNT16 | NA | WNT16 Entrez, Source | wingless-type MMTV integration site family, member 16 | 19151 | -0.239 | -0.0743 | No |
| 99 | NAT6 | NA | NAT6 Entrez, Source | N-acetyltransferase 6 | 19406 | -0.261 | -0.0815 | No |
| 100 | ADAM15 | NA | ADAM15 Entrez, Source | ADAM metallopeptidase domain 15 (metargidin) | 19419 | -0.262 | -0.0799 | No |
| 101 | PYCRL | NA | PYCRL Entrez, Source | pyrroline-5-carboxylate reductase-like | 19876 | -0.308 | -0.0942 | No |
| 102 | PTGIR | NA | PTGIR Entrez, Source | prostaglandin I2 (prostacyclin) receptor (IP) | 19950 | -0.315 | -0.0943 | No |
| 103 | RPL13P5 | NA | 20215 | -0.343 | -0.1013 | No | ||
| 104 | SLC3A1 | NA | SLC3A1 Entrez, Source | 1 | 20428 | -0.371 | -0.1061 | No |
| 105 | ZBTB22 | NA | ZBTB22 Entrez, Source | zinc finger and BTB domain containing 22 | 20712 | -0.408 | -0.1132 | No |
| 106 | CNTF | NA | CNTF Entrez, Source | ciliary neurotrophic factor | 20787 | -0.420 | -0.1125 | No |
| 107 | ABLIM2 | NA | ABLIM2 Entrez, Source | actin binding LIM protein family, member 2 | 21583 | -0.549 | -0.1373 | No |
| 108 | EDEM1 | NA | EDEM1 Entrez, Source | ER degradation enhancer, mannosidase alpha-like 1 | 21612 | -0.554 | -0.1339 | No |
| 109 | MORN1 | NA | MORN1 Entrez, Source | MORN repeat containing 1 | 21622 | -0.555 | -0.1298 | No |
| 110 | ZSCAN2 | NA | ZSCAN2 Entrez, Source | zinc finger and SCAN domain containing 2 | 21832 | -0.597 | -0.1327 | No |
| 111 | CD7 | NA | CD7 Entrez, Source | CD7 molecule | 22149 | -0.656 | -0.1390 | No |
| 112 | TGM1 | NA | TGM1 Entrez, Source | transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) | 22653 | -0.766 | -0.1514 | No |
| 113 | C21orf96 | NA | C21ORF96 Entrez, Source | chromosome 21 open reading frame 96 | 23191 | -0.876 | -0.1641 | No |
| 114 | FOXRED2 | NA | FOXRED2 Entrez, Source | FAD-dependent oxidoreductase domain containing 2 | 23406 | -0.927 | -0.1645 | No |
| 115 | CASP5 | NA | CASP5 Entrez, Source | caspase 5, apoptosis-related cysteine peptidase | 23451 | -0.937 | -0.1586 | No |
| 116 | PI15 | NA | PI15 Entrez, Source | peptidase inhibitor 15 | 23943 | -1.075 | -0.1680 | No |
| 117 | METTL10 | NA | 24217 | -1.155 | -0.1687 | No | ||
| 118 | MGA | NA | MGA Entrez, Source | MAX gene associated | 24635 | -1.287 | -0.1737 | No |
| 119 | CCNA1 | NA | CCNA1 Entrez, Source | cyclin A1 | 24811 | -1.349 | -0.1693 | No |
| 120 | HLA-DOB | NA | HLA-DOB Entrez, Source | major histocompatibility complex, class II, DO beta | 24993 | -1.416 | -0.1646 | No |
| 121 | NUPL1 | NA | NUPL1 Entrez, Source | nucleoporin like 1 | 25063 | -1.445 | -0.1555 | No |
| 122 | RNF125 | NA | RNF125 Entrez, Source | ring finger protein 125 | 25145 | -1.482 | -0.1466 | No |
| 123 | SACS | NA | SACS Entrez, Source | spastic ataxia of Charlevoix-Saguenay (sacsin) | 25688 | -1.735 | -0.1525 | No |
| 124 | RASGRF1 | NA | RASGRF1 Entrez, Source | Ras protein-specific guanine nucleotide-releasing factor 1 | 25761 | -1.774 | -0.1409 | No |
| 125 | CTF1 | NA | CTF1 Entrez, Source | cardiotrophin 1 | 26074 | -1.962 | -0.1366 | No |
| 126 | CLMN | NA | CLMN Entrez, Source | calmin (calponin-like, transmembrane) | 26191 | -2.052 | -0.1244 | No |
| 127 | FAM169A | NA | 26816 | -2.672 | -0.1259 | No | ||
| 128 | CARD10 | NA | CARD10 Entrez, Source | caspase recruitment domain family, member 10 | 26910 | -2.821 | -0.1066 | No |
| 129 | CHST15 | NA | 27108 | -3.403 | -0.0865 | No | ||
| 130 | SERPINB9 | NA | SERPINB9 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 27166 | -3.599 | -0.0597 | No |
| 131 | ECE2 | NA | ECE2 Entrez, Source | endothelin converting enzyme 2 | 27198 | -3.805 | -0.0302 | No |
| 132 | RUNX3 | NA | RUNX3 Entrez, Source | runt-related transcription factor 3 | 27278 | -4.352 | 0.0019 | No |