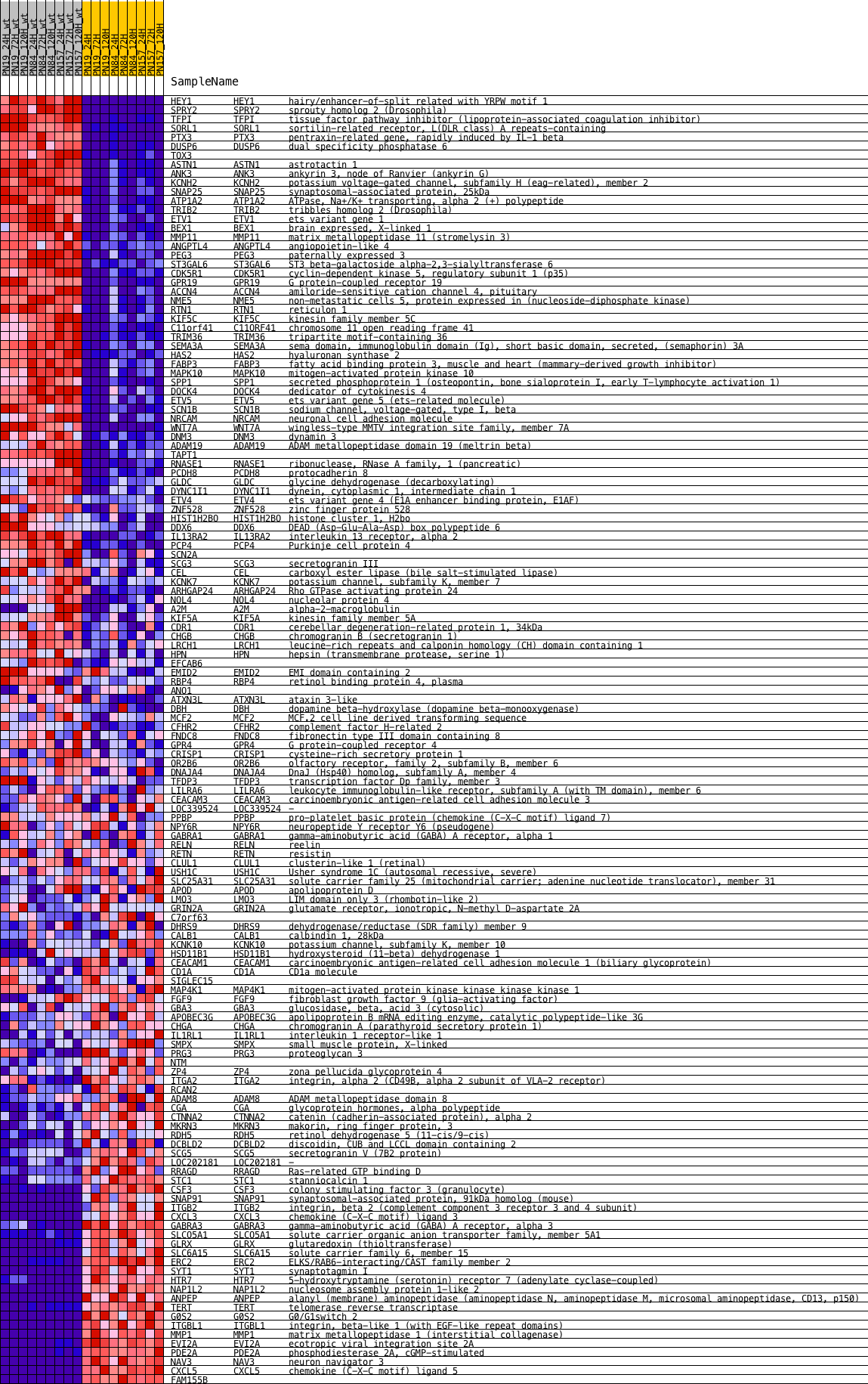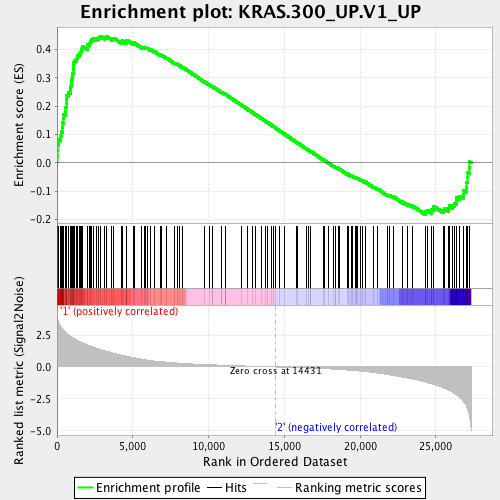
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | KRAS.300_UP.V1_UP |
| Enrichment Score (ES) | 0.44773784 |
| Normalized Enrichment Score (NES) | 1.7141532 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00402716 |
| FWER p-Value | 0.02 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HEY1 | NA | HEY1 Entrez, Source | hairy/enhancer-of-split related with YRPW motif 1 | 2 | 4.185 | 0.0233 | Yes |
| 2 | SPRY2 | NA | SPRY2 Entrez, Source | sprouty homolog 2 (Drosophila) | 9 | 3.989 | 0.0454 | Yes |
| 3 | TFPI | NA | TFPI Entrez, Source | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 43 | 3.706 | 0.0649 | Yes |
| 4 | SORL1 | NA | SORL1 Entrez, Source | sortilin-related receptor, L(DLR class) A repeats-containing | 119 | 3.420 | 0.0813 | Yes |
| 5 | PTX3 | NA | PTX3 Entrez, Source | pentraxin-related gene, rapidly induced by IL-1 beta | 227 | 3.226 | 0.0954 | Yes |
| 6 | DUSP6 | NA | DUSP6 Entrez, Source | dual specificity phosphatase 6 | 312 | 3.058 | 0.1094 | Yes |
| 7 | TOX3 | NA | 345 | 3.002 | 0.1250 | Yes | ||
| 8 | ASTN1 | NA | ASTN1 Entrez, Source | astrotactin 1 | 359 | 2.982 | 0.1412 | Yes |
| 9 | ANK3 | NA | ANK3 Entrez, Source | ankyrin 3, node of Ranvier (ankyrin G) | 399 | 2.930 | 0.1561 | Yes |
| 10 | KCNH2 | NA | KCNH2 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 2 | 426 | 2.897 | 0.1714 | Yes |
| 11 | SNAP25 | NA | SNAP25 Entrez, Source | synaptosomal-associated protein, 25kDa | 546 | 2.722 | 0.1822 | Yes |
| 12 | ATP1A2 | NA | ATP1A2 Entrez, Source | ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide | 579 | 2.691 | 0.1961 | Yes |
| 13 | TRIB2 | NA | TRIB2 Entrez, Source | tribbles homolog 2 (Drosophila) | 607 | 2.668 | 0.2100 | Yes |
| 14 | ETV1 | NA | ETV1 Entrez, Source | ets variant gene 1 | 615 | 2.660 | 0.2246 | Yes |
| 15 | BEX1 | NA | BEX1 Entrez, Source | brain expressed, X-linked 1 | 645 | 2.626 | 0.2382 | Yes |
| 16 | MMP11 | NA | MMP11 Entrez, Source | matrix metallopeptidase 11 (stromelysin 3) | 736 | 2.539 | 0.2491 | Yes |
| 17 | ANGPTL4 | NA | ANGPTL4 Entrez, Source | angiopoietin-like 4 | 866 | 2.417 | 0.2579 | Yes |
| 18 | PEG3 | NA | PEG3 Entrez, Source | paternally expressed 3 | 886 | 2.400 | 0.2706 | Yes |
| 19 | ST3GAL6 | NA | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 923 | 2.370 | 0.2825 | Yes |
| 20 | CDK5R1 | NA | CDK5R1 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 965 | 2.340 | 0.2941 | Yes |
| 21 | GPR19 | NA | GPR19 Entrez, Source | G protein-coupled receptor 19 | 1003 | 2.316 | 0.3057 | Yes |
| 22 | ACCN4 | NA | ACCN4 Entrez, Source | amiloride-sensitive cation channel 4, pituitary | 1041 | 2.288 | 0.3171 | Yes |
| 23 | NME5 | NA | NME5 Entrez, Source | non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase) | 1066 | 2.268 | 0.3289 | Yes |
| 24 | RTN1 | NA | RTN1 Entrez, Source | reticulon 1 | 1084 | 2.254 | 0.3409 | Yes |
| 25 | KIF5C | NA | KIF5C Entrez, Source | kinesin family member 5C | 1092 | 2.248 | 0.3532 | Yes |
| 26 | C11orf41 | NA | C11ORF41 Entrez, Source | chromosome 11 open reading frame 41 | 1164 | 2.200 | 0.3629 | Yes |
| 27 | TRIM36 | NA | TRIM36 Entrez, Source | tripartite motif-containing 36 | 1308 | 2.095 | 0.3693 | Yes |
| 28 | SEMA3A | NA | SEMA3A Entrez, Source | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | 1323 | 2.086 | 0.3805 | Yes |
| 29 | HAS2 | NA | HAS2 Entrez, Source | hyaluronan synthase 2 | 1487 | 1.986 | 0.3856 | Yes |
| 30 | FABP3 | NA | FABP3 Entrez, Source | fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 1573 | 1.939 | 0.3933 | Yes |
| 31 | MAPK10 | NA | MAPK10 Entrez, Source | mitogen-activated protein kinase 10 | 1618 | 1.911 | 0.4024 | Yes |
| 32 | SPP1 | NA | SPP1 Entrez, Source | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 1647 | 1.897 | 0.4119 | Yes |
| 33 | DOCK4 | NA | DOCK4 Entrez, Source | dedicator of cytokinesis 4 | 1989 | 1.731 | 0.4091 | Yes |
| 34 | ETV5 | NA | ETV5 Entrez, Source | ets variant gene 5 (ets-related molecule) | 1991 | 1.731 | 0.4187 | Yes |
| 35 | SCN1B | NA | SCN1B Entrez, Source | sodium channel, voltage-gated, type I, beta | 2152 | 1.654 | 0.4221 | Yes |
| 36 | NRCAM | NA | NRCAM Entrez, Source | neuronal cell adhesion molecule | 2208 | 1.630 | 0.4292 | Yes |
| 37 | WNT7A | NA | WNT7A Entrez, Source | wingless-type MMTV integration site family, member 7A | 2246 | 1.618 | 0.4368 | Yes |
| 38 | DNM3 | NA | DNM3 Entrez, Source | dynamin 3 | 2393 | 1.551 | 0.4401 | Yes |
| 39 | ADAM19 | NA | ADAM19 Entrez, Source | ADAM metallopeptidase domain 19 (meltrin beta) | 2593 | 1.464 | 0.4410 | Yes |
| 40 | TAPT1 | NA | 2742 | 1.403 | 0.4434 | Yes | ||
| 41 | RNASE1 | NA | RNASE1 Entrez, Source | ribonuclease, RNase A family, 1 (pancreatic) | 2834 | 1.373 | 0.4477 | Yes |
| 42 | PCDH8 | NA | PCDH8 Entrez, Source | protocadherin 8 | 3119 | 1.269 | 0.4444 | No |
| 43 | GLDC | NA | GLDC Entrez, Source | glycine dehydrogenase (decarboxylating) | 3266 | 1.223 | 0.4458 | No |
| 44 | DYNC1I1 | NA | DYNC1I1 Entrez, Source | dynein, cytoplasmic 1, intermediate chain 1 | 3602 | 1.115 | 0.4398 | No |
| 45 | ETV4 | NA | ETV4 Entrez, Source | ets variant gene 4 (E1A enhancer binding protein, E1AF) | 3740 | 1.071 | 0.4407 | No |
| 46 | ZNF528 | NA | ZNF528 Entrez, Source | zinc finger protein 528 | 4241 | 0.922 | 0.4275 | No |
| 47 | HIST1H2BO | NA | HIST1H2BO Entrez, Source | histone cluster 1, H2bo | 4289 | 0.908 | 0.4308 | No |
| 48 | DDX6 | NA | DDX6 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | 4551 | 0.850 | 0.4260 | No |
| 49 | IL13RA2 | NA | IL13RA2 Entrez, Source | interleukin 13 receptor, alpha 2 | 4557 | 0.849 | 0.4305 | No |
| 50 | PCP4 | NA | PCP4 Entrez, Source | Purkinje cell protein 4 | 4601 | 0.842 | 0.4337 | No |
| 51 | SCN2A | NA | 5072 | 0.728 | 0.4205 | No | ||
| 52 | SCG3 | NA | SCG3 Entrez, Source | secretogranin III | 5082 | 0.725 | 0.4242 | No |
| 53 | CEL | NA | CEL Entrez, Source | carboxyl ester lipase (bile salt-stimulated lipase) | 5598 | 0.597 | 0.4086 | No |
| 54 | KCNK7 | NA | KCNK7 Entrez, Source | potassium channel, subfamily K, member 7 | 5761 | 0.568 | 0.4058 | No |
| 55 | ARHGAP24 | NA | ARHGAP24 Entrez, Source | Rho GTPase activating protein 24 | 5839 | 0.555 | 0.4061 | No |
| 56 | NOL4 | NA | NOL4 Entrez, Source | nucleolar protein 4 | 5940 | 0.539 | 0.4054 | No |
| 57 | A2M | NA | A2M Entrez, Source | alpha-2-macroglobulin | 6145 | 0.506 | 0.4007 | No |
| 58 | KIF5A | NA | KIF5A Entrez, Source | kinesin family member 5A | 6447 | 0.458 | 0.3922 | No |
| 59 | CDR1 | NA | CDR1 Entrez, Source | cerebellar degeneration-related protein 1, 34kDa | 6804 | 0.418 | 0.3814 | No |
| 60 | CHGB | NA | CHGB Entrez, Source | chromogranin B (secretogranin 1) | 6891 | 0.409 | 0.3806 | No |
| 61 | LRCH1 | NA | LRCH1 Entrez, Source | leucine-rich repeats and calponin homology (CH) domain containing 1 | 7246 | 0.371 | 0.3696 | No |
| 62 | HPN | NA | HPN Entrez, Source | hepsin (transmembrane protease, serine 1) | 7770 | 0.321 | 0.3522 | No |
| 63 | EFCAB6 | NA | 7923 | 0.307 | 0.3483 | No | ||
| 64 | EMID2 | NA | EMID2 Entrez, Source | EMI domain containing 2 | 8068 | 0.295 | 0.3447 | No |
| 65 | RBP4 | NA | RBP4 Entrez, Source | retinol binding protein 4, plasma | 8284 | 0.278 | 0.3383 | No |
| 66 | ANO1 | NA | 9757 | 0.190 | 0.2852 | No | ||
| 67 | ATXN3L | NA | ATXN3L Entrez, Source | ataxin 3-like | 10031 | 0.177 | 0.2762 | No |
| 68 | DBH | NA | DBH Entrez, Source | dopamine beta-hydroxylase (dopamine beta-monooxygenase) | 10232 | 0.168 | 0.2698 | No |
| 69 | MCF2 | NA | MCF2 Entrez, Source | MCF.2 cell line derived transforming sequence | 10285 | 0.166 | 0.2688 | No |
| 70 | CFHR2 | NA | CFHR2 Entrez, Source | complement factor H-related 2 | 10851 | 0.139 | 0.2488 | No |
| 71 | FNDC8 | NA | FNDC8 Entrez, Source | fibronectin type III domain containing 8 | 10863 | 0.139 | 0.2491 | No |
| 72 | GPR4 | NA | GPR4 Entrez, Source | G protein-coupled receptor 4 | 11084 | 0.128 | 0.2418 | No |
| 73 | CRISP1 | NA | CRISP1 Entrez, Source | cysteine-rich secretory protein 1 | 11103 | 0.127 | 0.2418 | No |
| 74 | OR2B6 | NA | OR2B6 Entrez, Source | olfactory receptor, family 2, subfamily B, member 6 | 11131 | 0.126 | 0.2415 | No |
| 75 | DNAJA4 | NA | DNAJA4 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 4 | 12196 | 0.083 | 0.2029 | No |
| 76 | TFDP3 | NA | TFDP3 Entrez, Source | transcription factor Dp family, member 3 | 12565 | 0.069 | 0.1897 | No |
| 77 | LILRA6 | NA | LILRA6 Entrez, Source | leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 | 12898 | 0.057 | 0.1778 | No |
| 78 | CEACAM3 | NA | CEACAM3 Entrez, Source | carcinoembryonic antigen-related cell adhesion molecule 3 | 13095 | 0.049 | 0.1709 | No |
| 79 | LOC339524 | NA | LOC339524 Entrez, Source | - | 13467 | 0.035 | 0.1574 | No |
| 80 | PPBP | NA | PPBP Entrez, Source | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | 13740 | 0.024 | 0.1476 | No |
| 81 | NPY6R | NA | NPY6R Entrez, Source | neuropeptide Y receptor Y6 (pseudogene) | 13898 | 0.018 | 0.1419 | No |
| 82 | GABRA1 | NA | GABRA1 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 1 | 14115 | 0.011 | 0.1340 | No |
| 83 | RELN | NA | RELN Entrez, Source | reelin | 14279 | 0.005 | 0.1280 | No |
| 84 | RETN | NA | RETN Entrez, Source | resistin | 14399 | 0.001 | 0.1237 | No |
| 85 | CLUL1 | NA | CLUL1 Entrez, Source | clusterin-like 1 (retinal) | 14663 | -0.008 | 0.1140 | No |
| 86 | USH1C | NA | USH1C Entrez, Source | Usher syndrome 1C (autosomal recessive, severe) | 14989 | -0.021 | 0.1022 | No |
| 87 | SLC25A31 | NA | SLC25A31 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 | 15810 | -0.052 | 0.0723 | No |
| 88 | APOD | NA | APOD Entrez, Source | apolipoprotein D | 15857 | -0.054 | 0.0709 | No |
| 89 | LMO3 | NA | LMO3 Entrez, Source | LIM domain only 3 (rhombotin-like 2) | 16466 | -0.080 | 0.0490 | No |
| 90 | GRIN2A | NA | GRIN2A Entrez, Source | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 16591 | -0.084 | 0.0449 | No |
| 91 | C7orf63 | NA | 16691 | -0.089 | 0.0418 | No | ||
| 92 | DHRS9 | NA | DHRS9 Entrez, Source | dehydrogenase/reductase (SDR family) member 9 | 16733 | -0.090 | 0.0408 | No |
| 93 | CALB1 | NA | CALB1 Entrez, Source | calbindin 1, 28kDa | 17578 | -0.128 | 0.0105 | No |
| 94 | KCNK10 | NA | KCNK10 Entrez, Source | potassium channel, subfamily K, member 10 | 17651 | -0.132 | 0.0086 | No |
| 95 | HSD11B1 | NA | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 17896 | -0.147 | 0.0004 | No |
| 96 | CEACAM1 | NA | CEACAM1 Entrez, Source | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 18265 | -0.170 | -0.0122 | No |
| 97 | CD1A | NA | CD1A Entrez, Source | CD1a molecule | 18355 | -0.175 | -0.0145 | No |
| 98 | SIGLEC15 | NA | 18395 | -0.178 | -0.0149 | No | ||
| 99 | MAP4K1 | NA | MAP4K1 Entrez, Source | mitogen-activated protein kinase kinase kinase kinase 1 | 18558 | -0.189 | -0.0198 | No |
| 100 | FGF9 | NA | FGF9 Entrez, Source | fibroblast growth factor 9 (glia-activating factor) | 18634 | -0.196 | -0.0215 | No |
| 101 | GBA3 | NA | GBA3 Entrez, Source | glucosidase, beta, acid 3 (cytosolic) | 19138 | -0.238 | -0.0387 | No |
| 102 | APOBEC3G | NA | APOBEC3G Entrez, Source | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G | 19256 | -0.248 | -0.0416 | No |
| 103 | CHGA | NA | CHGA Entrez, Source | chromogranin A (parathyroid secretory protein 1) | 19395 | -0.260 | -0.0452 | No |
| 104 | IL1RL1 | NA | IL1RL1 Entrez, Source | interleukin 1 receptor-like 1 | 19520 | -0.272 | -0.0482 | No |
| 105 | SMPX | NA | SMPX Entrez, Source | small muscle protein, X-linked | 19685 | -0.288 | -0.0527 | No |
| 106 | PRG3 | NA | PRG3 Entrez, Source | proteoglycan 3 | 19742 | -0.294 | -0.0531 | No |
| 107 | NTM | NA | 19833 | -0.303 | -0.0547 | No | ||
| 108 | ZP4 | NA | ZP4 Entrez, Source | zona pellucida glycoprotein 4 | 20019 | -0.322 | -0.0597 | No |
| 109 | ITGA2 | NA | ITGA2 Entrez, Source | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 20150 | -0.337 | -0.0626 | No |
| 110 | RCAN2 | NA | 20320 | -0.358 | -0.0668 | No | ||
| 111 | ADAM8 | NA | ADAM8 Entrez, Source | ADAM metallopeptidase domain 8 | 20906 | -0.438 | -0.0859 | No |
| 112 | CGA | NA | CGA Entrez, Source | glycoprotein hormones, alpha polypeptide | 21170 | -0.477 | -0.0929 | No |
| 113 | CTNNA2 | NA | CTNNA2 Entrez, Source | catenin (cadherin-associated protein), alpha 2 | 21819 | -0.595 | -0.1134 | No |
| 114 | MKRN3 | NA | MKRN3 Entrez, Source | makorin, ring finger protein, 3 | 21929 | -0.614 | -0.1140 | No |
| 115 | RDH5 | NA | RDH5 Entrez, Source | retinol dehydrogenase 5 (11-cis/9-cis) | 22170 | -0.660 | -0.1191 | No |
| 116 | DCBLD2 | NA | DCBLD2 Entrez, Source | discoidin, CUB and LCCL domain containing 2 | 22793 | -0.793 | -0.1375 | No |
| 117 | SCG5 | NA | SCG5 Entrez, Source | secretogranin V (7B2 protein) | 23141 | -0.867 | -0.1455 | No |
| 118 | LOC202181 | NA | LOC202181 Entrez, Source | - | 23425 | -0.932 | -0.1507 | No |
| 119 | RRAGD | NA | RRAGD Entrez, Source | Ras-related GTP binding D | 24282 | -1.175 | -0.1756 | No |
| 120 | STC1 | NA | STC1 Entrez, Source | stanniocalcin 1 | 24330 | -1.189 | -0.1707 | No |
| 121 | CSF3 | NA | CSF3 Entrez, Source | colony stimulating factor 3 (granulocyte) | 24446 | -1.225 | -0.1680 | No |
| 122 | SNAP91 | NA | SNAP91 Entrez, Source | synaptosomal-associated protein, 91kDa homolog (mouse) | 24696 | -1.307 | -0.1699 | No |
| 123 | ITGB2 | NA | ITGB2 Entrez, Source | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 24721 | -1.317 | -0.1634 | No |
| 124 | CXCL3 | NA | CXCL3 Entrez, Source | chemokine (C-X-C motif) ligand 3 | 24820 | -1.350 | -0.1595 | No |
| 125 | GABRA3 | NA | GABRA3 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | 24833 | -1.355 | -0.1523 | No |
| 126 | SLCO5A1 | NA | SLCO5A1 Entrez, Source | solute carrier organic anion transporter family, member 5A1 | 25484 | -1.634 | -0.1671 | No |
| 127 | GLRX | NA | GLRX Entrez, Source | glutaredoxin (thioltransferase) | 25564 | -1.671 | -0.1607 | No |
| 128 | SLC6A15 | NA | SLC6A15 Entrez, Source | solute carrier family 6, member 15 | 25846 | -1.821 | -0.1608 | No |
| 129 | ERC2 | NA | ERC2 Entrez, Source | ELKS/RAB6-interacting/CAST family member 2 | 25870 | -1.839 | -0.1514 | No |
| 130 | SYT1 | NA | SYT1 Entrez, Source | synaptotagmin I | 26109 | -1.987 | -0.1490 | No |
| 131 | HTR7 | NA | HTR7 Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled) | 26240 | -2.081 | -0.1422 | No |
| 132 | NAP1L2 | NA | NAP1L2 Entrez, Source | nucleosome assembly protein 1-like 2 | 26330 | -2.152 | -0.1334 | No |
| 133 | ANPEP | NA | ANPEP Entrez, Source | alanyl (membrane) aminopeptidase (aminopeptidase N, aminopeptidase M, microsomal aminopeptidase, CD13, p150) | 26355 | -2.176 | -0.1221 | No |
| 134 | TERT | NA | TERT Entrez, Source | telomerase reverse transcriptase | 26573 | -2.357 | -0.1169 | No |
| 135 | G0S2 | NA | G0S2 Entrez, Source | G0/G1switch 2 | 26789 | -2.628 | -0.1102 | No |
| 136 | ITGBL1 | NA | ITGBL1 Entrez, Source | integrin, beta-like 1 (with EGF-like repeat domains) | 26841 | -2.730 | -0.0968 | No |
| 137 | MMP1 | NA | MMP1 Entrez, Source | matrix metallopeptidase 1 (interstitial collagenase) | 27012 | -3.065 | -0.0859 | No |
| 138 | EVI2A | NA | EVI2A Entrez, Source | ecotropic viral integration site 2A | 27034 | -3.146 | -0.0691 | No |
| 139 | PDE2A | NA | PDE2A Entrez, Source | phosphodiesterase 2A, cGMP-stimulated | 27080 | -3.285 | -0.0524 | No |
| 140 | NAV3 | NA | NAV3 Entrez, Source | neuron navigator 3 | 27085 | -3.304 | -0.0340 | No |
| 141 | CXCL5 | NA | CXCL5 Entrez, Source | chemokine (C-X-C motif) ligand 5 | 27183 | -3.696 | -0.0169 | No |
| 142 | FAM155B | NA | 27228 | -3.985 | 0.0037 | No |