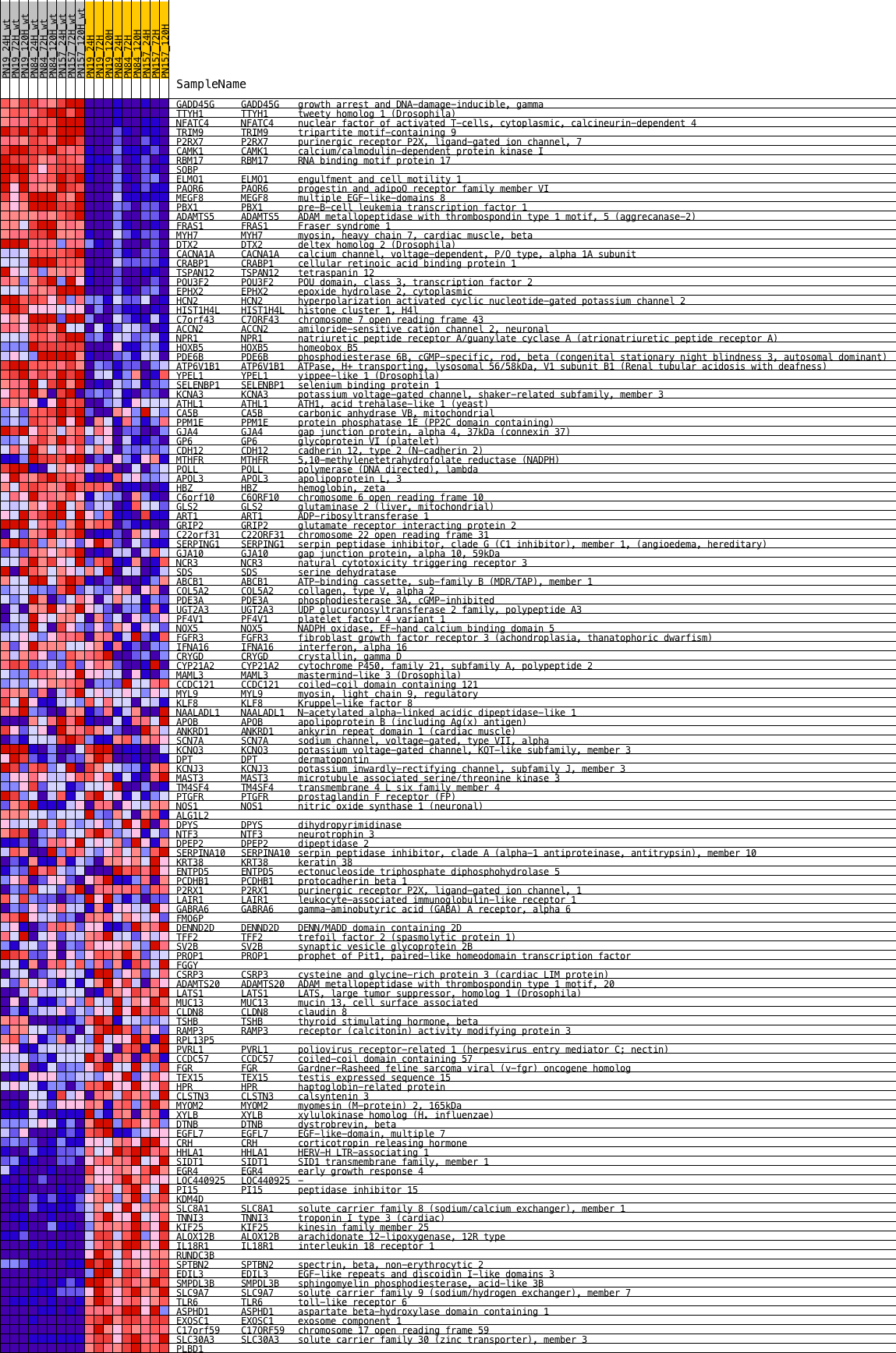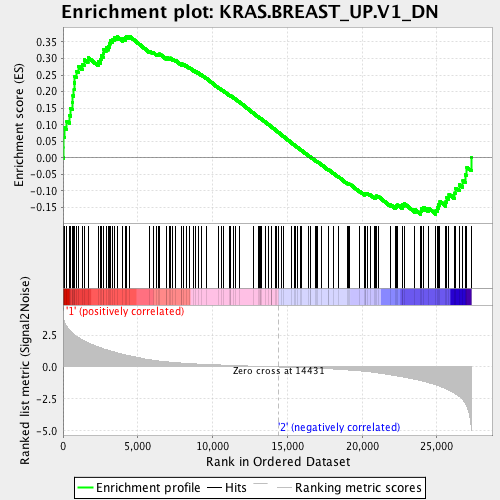
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | KRAS.BREAST_UP.V1_DN |
| Enrichment Score (ES) | 0.3675649 |
| Normalized Enrichment Score (NES) | 1.6177967 |
| Nominal p-value | 0.0019880715 |
| FDR q-value | 0.009711223 |
| FWER p-Value | 0.08 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GADD45G | NA | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 25 | 3.828 | 0.0311 | Yes |
| 2 | TTYH1 | NA | TTYH1 Entrez, Source | tweety homolog 1 (Drosophila) | 27 | 3.791 | 0.0627 | Yes |
| 3 | NFATC4 | NA | NFATC4 Entrez, Source | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | 78 | 3.543 | 0.0905 | Yes |
| 4 | TRIM9 | NA | TRIM9 Entrez, Source | tripartite motif-containing 9 | 251 | 3.172 | 0.1107 | Yes |
| 5 | P2RX7 | NA | P2RX7 Entrez, Source | purinergic receptor P2X, ligand-gated ion channel, 7 | 416 | 2.906 | 0.1290 | Yes |
| 6 | CAMK1 | NA | CAMK1 Entrez, Source | calcium/calmodulin-dependent protein kinase I | 505 | 2.787 | 0.1490 | Yes |
| 7 | RBM17 | NA | RBM17 Entrez, Source | RNA binding motif protein 17 | 610 | 2.667 | 0.1675 | Yes |
| 8 | SOBP | NA | 657 | 2.617 | 0.1877 | Yes | ||
| 9 | ELMO1 | NA | ELMO1 Entrez, Source | engulfment and cell motility 1 | 727 | 2.544 | 0.2064 | Yes |
| 10 | PAQR6 | NA | PAQR6 Entrez, Source | progestin and adipoQ receptor family member VI | 750 | 2.521 | 0.2267 | Yes |
| 11 | MEGF8 | NA | MEGF8 Entrez, Source | multiple EGF-like-domains 8 | 786 | 2.489 | 0.2462 | Yes |
| 12 | PBX1 | NA | PBX1 Entrez, Source | pre-B-cell leukemia transcription factor 1 | 894 | 2.392 | 0.2622 | Yes |
| 13 | ADAMTS5 | NA | ADAMTS5 Entrez, Source | ADAM metallopeptidase with thrombospondin type 1 motif, 5 (aggrecanase-2) | 1050 | 2.278 | 0.2756 | Yes |
| 14 | FRAS1 | NA | FRAS1 Entrez, Source | Fraser syndrome 1 | 1318 | 2.089 | 0.2832 | Yes |
| 15 | MYH7 | NA | MYH7 Entrez, Source | myosin, heavy chain 7, cardiac muscle, beta | 1433 | 2.014 | 0.2959 | Yes |
| 16 | DTX2 | NA | DTX2 Entrez, Source | deltex homolog 2 (Drosophila) | 1667 | 1.889 | 0.3031 | Yes |
| 17 | CACNA1A | NA | CACNA1A Entrez, Source | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | 2344 | 1.578 | 0.2914 | Yes |
| 18 | CRABP1 | NA | CRABP1 Entrez, Source | cellular retinoic acid binding protein 1 | 2500 | 1.506 | 0.2983 | Yes |
| 19 | TSPAN12 | NA | TSPAN12 Entrez, Source | tetraspanin 12 | 2565 | 1.475 | 0.3083 | Yes |
| 20 | POU3F2 | NA | POU3F2 Entrez, Source | POU domain, class 3, transcription factor 2 | 2684 | 1.428 | 0.3159 | Yes |
| 21 | EPHX2 | NA | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 2718 | 1.416 | 0.3265 | Yes |
| 22 | HCN2 | NA | HCN2 Entrez, Source | hyperpolarization activated cyclic nucleotide-gated potassium channel 2 | 2874 | 1.359 | 0.3322 | Yes |
| 23 | HIST1H4L | NA | HIST1H4L Entrez, Source | histone cluster 1, H4l | 3054 | 1.292 | 0.3364 | Yes |
| 24 | C7orf43 | NA | C7ORF43 Entrez, Source | chromosome 7 open reading frame 43 | 3121 | 1.268 | 0.3446 | Yes |
| 25 | ACCN2 | NA | ACCN2 Entrez, Source | amiloride-sensitive cation channel 2, neuronal | 3145 | 1.259 | 0.3542 | Yes |
| 26 | NPR1 | NA | NPR1 Entrez, Source | natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) | 3326 | 1.207 | 0.3577 | Yes |
| 27 | HOXB5 | NA | HOXB5 Entrez, Source | homeobox B5 | 3457 | 1.167 | 0.3627 | Yes |
| 28 | PDE6B | NA | PDE6B Entrez, Source | phosphodiesterase 6B, cGMP-specific, rod, beta (congenital stationary night blindness 3, autosomal dominant) | 3624 | 1.108 | 0.3658 | Yes |
| 29 | ATP6V1B1 | NA | ATP6V1B1 Entrez, Source | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 (Renal tubular acidosis with deafness) | 4005 | 0.986 | 0.3601 | Yes |
| 30 | YPEL1 | NA | YPEL1 Entrez, Source | yippee-like 1 (Drosophila) | 4152 | 0.945 | 0.3626 | Yes |
| 31 | SELENBP1 | NA | SELENBP1 Entrez, Source | selenium binding protein 1 | 4235 | 0.924 | 0.3673 | Yes |
| 32 | KCNA3 | NA | KCNA3 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 3 | 4429 | 0.877 | 0.3676 | Yes |
| 33 | ATHL1 | NA | ATHL1 Entrez, Source | ATH1, acid trehalase-like 1 (yeast) | 5782 | 0.563 | 0.3226 | No |
| 34 | CA5B | NA | CA5B Entrez, Source | carbonic anhydrase VB, mitochondrial | 6013 | 0.524 | 0.3185 | No |
| 35 | PPM1E | NA | PPM1E Entrez, Source | protein phosphatase 1E (PP2C domain containing) | 6260 | 0.488 | 0.3135 | No |
| 36 | GJA4 | NA | GJA4 Entrez, Source | gap junction protein, alpha 4, 37kDa (connexin 37) | 6412 | 0.464 | 0.3118 | No |
| 37 | GP6 | NA | GP6 Entrez, Source | glycoprotein VI (platelet) | 6465 | 0.456 | 0.3137 | No |
| 38 | CDH12 | NA | CDH12 Entrez, Source | cadherin 12, type 2 (N-cadherin 2) | 6932 | 0.404 | 0.3000 | No |
| 39 | MTHFR | NA | MTHFR Entrez, Source | 5,10-methylenetetrahydrofolate reductase (NADPH) | 6944 | 0.403 | 0.3029 | No |
| 40 | POLL | NA | POLL Entrez, Source | polymerase (DNA directed), lambda | 7088 | 0.388 | 0.3009 | No |
| 41 | APOL3 | NA | APOL3 Entrez, Source | apolipoprotein L, 3 | 7159 | 0.381 | 0.3015 | No |
| 42 | HBZ | NA | HBZ Entrez, Source | hemoglobin, zeta | 7336 | 0.361 | 0.2981 | No |
| 43 | C6orf10 | NA | C6ORF10 Entrez, Source | chromosome 6 open reading frame 10 | 7528 | 0.342 | 0.2939 | No |
| 44 | GLS2 | NA | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 7913 | 0.308 | 0.2824 | No |
| 45 | ART1 | NA | ART1 Entrez, Source | ADP-ribosyltransferase 1 | 7921 | 0.308 | 0.2847 | No |
| 46 | GRIP2 | NA | GRIP2 Entrez, Source | glutamate receptor interacting protein 2 | 8064 | 0.295 | 0.2819 | No |
| 47 | C22orf31 | NA | C22ORF31 Entrez, Source | chromosome 22 open reading frame 31 | 8242 | 0.282 | 0.2778 | No |
| 48 | SERPING1 | NA | SERPING1 Entrez, Source | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) | 8450 | 0.266 | 0.2724 | No |
| 49 | GJA10 | NA | GJA10 Entrez, Source | gap junction protein, alpha 10, 59kDa | 8754 | 0.244 | 0.2633 | No |
| 50 | NCR3 | NA | NCR3 Entrez, Source | natural cytotoxicity triggering receptor 3 | 8890 | 0.237 | 0.2603 | No |
| 51 | SDS | NA | SDS Entrez, Source | serine dehydratase | 9038 | 0.228 | 0.2568 | No |
| 52 | ABCB1 | NA | ABCB1 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 9255 | 0.216 | 0.2507 | No |
| 53 | COL5A2 | NA | COL5A2 Entrez, Source | collagen, type V, alpha 2 | 9563 | 0.200 | 0.2411 | No |
| 54 | PDE3A | NA | PDE3A Entrez, Source | phosphodiesterase 3A, cGMP-inhibited | 10425 | 0.159 | 0.2107 | No |
| 55 | UGT2A3 | NA | UGT2A3 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide A3 | 10580 | 0.152 | 0.2063 | No |
| 56 | PF4V1 | NA | PF4V1 Entrez, Source | platelet factor 4 variant 1 | 10717 | 0.145 | 0.2025 | No |
| 57 | NOX5 | NA | NOX5 Entrez, Source | NADPH oxidase, EF-hand calcium binding domain 5 | 11143 | 0.126 | 0.1880 | No |
| 58 | FGFR3 | NA | FGFR3 Entrez, Source | fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) | 11229 | 0.122 | 0.1859 | No |
| 59 | IFNA16 | NA | IFNA16 Entrez, Source | interferon, alpha 16 | 11230 | 0.122 | 0.1869 | No |
| 60 | CRYGD | NA | CRYGD Entrez, Source | crystallin, gamma D | 11380 | 0.114 | 0.1824 | No |
| 61 | CYP21A2 | NA | CYP21A2 Entrez, Source | cytochrome P450, family 21, subfamily A, polypeptide 2 | 11564 | 0.108 | 0.1765 | No |
| 62 | MAML3 | NA | MAML3 Entrez, Source | mastermind-like 3 (Drosophila) | 11786 | 0.098 | 0.1692 | No |
| 63 | CCDC121 | NA | CCDC121 Entrez, Source | coiled-coil domain containing 121 | 12714 | 0.063 | 0.1357 | No |
| 64 | MYL9 | NA | MYL9 Entrez, Source | myosin, light chain 9, regulatory | 13092 | 0.049 | 0.1222 | No |
| 65 | KLF8 | NA | KLF8 Entrez, Source | Kruppel-like factor 8 | 13110 | 0.049 | 0.1220 | No |
| 66 | NAALADL1 | NA | NAALADL1 Entrez, Source | N-acetylated alpha-linked acidic dipeptidase-like 1 | 13199 | 0.045 | 0.1191 | No |
| 67 | APOB | NA | APOB Entrez, Source | apolipoprotein B (including Ag(x) antigen) | 13212 | 0.045 | 0.1191 | No |
| 68 | ANKRD1 | NA | ANKRD1 Entrez, Source | ankyrin repeat domain 1 (cardiac muscle) | 13268 | 0.042 | 0.1174 | No |
| 69 | SCN7A | NA | SCN7A Entrez, Source | sodium channel, voltage-gated, type VII, alpha | 13567 | 0.030 | 0.1067 | No |
| 70 | KCNQ3 | NA | KCNQ3 Entrez, Source | potassium voltage-gated channel, KQT-like subfamily, member 3 | 13718 | 0.025 | 0.1014 | No |
| 71 | DPT | NA | DPT Entrez, Source | dermatopontin | 13914 | 0.018 | 0.0944 | No |
| 72 | KCNJ3 | NA | KCNJ3 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 3 | 13915 | 0.018 | 0.0945 | No |
| 73 | MAST3 | NA | MAST3 Entrez, Source | microtubule associated serine/threonine kinase 3 | 13974 | 0.016 | 0.0925 | No |
| 74 | TM4SF4 | NA | TM4SF4 Entrez, Source | transmembrane 4 L six family member 4 | 14180 | 0.009 | 0.0851 | No |
| 75 | PTGFR | NA | PTGFR Entrez, Source | prostaglandin F receptor (FP) | 14270 | 0.005 | 0.0818 | No |
| 76 | NOS1 | NA | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 14403 | 0.001 | 0.0770 | No |
| 77 | ALG1L2 | NA | 14604 | -0.006 | 0.0697 | No | ||
| 78 | DPYS | NA | DPYS Entrez, Source | dihydropyrimidinase | 14745 | -0.011 | 0.0646 | No |
| 79 | NTF3 | NA | NTF3 Entrez, Source | neurotrophin 3 | 15279 | -0.032 | 0.0453 | No |
| 80 | DPEP2 | NA | DPEP2 Entrez, Source | dipeptidase 2 | 15503 | -0.041 | 0.0374 | No |
| 81 | SERPINA10 | NA | SERPINA10 Entrez, Source | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 | 15534 | -0.042 | 0.0367 | No |
| 82 | KRT38 | NA | KRT38 Entrez, Source | keratin 38 | 15700 | -0.048 | 0.0310 | No |
| 83 | ENTPD5 | NA | ENTPD5 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 5 | 15853 | -0.054 | 0.0259 | No |
| 84 | PCDHB1 | NA | PCDHB1 Entrez, Source | protocadherin beta 1 | 15980 | -0.058 | 0.0217 | No |
| 85 | P2RX1 | NA | P2RX1 Entrez, Source | purinergic receptor P2X, ligand-gated ion channel, 1 | 16433 | -0.079 | 0.0058 | No |
| 86 | LAIR1 | NA | LAIR1 Entrez, Source | leukocyte-associated immunoglobulin-like receptor 1 | 16521 | -0.082 | 0.0033 | No |
| 87 | GABRA6 | NA | GABRA6 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 6 | 16571 | -0.084 | 0.0022 | No |
| 88 | FMO6P | NA | 16855 | -0.096 | -0.0074 | No | ||
| 89 | DENND2D | NA | DENND2D Entrez, Source | DENN/MADD domain containing 2D | 16954 | -0.099 | -0.0102 | No |
| 90 | TFF2 | NA | TFF2 Entrez, Source | trefoil factor 2 (spasmolytic protein 1) | 17043 | -0.103 | -0.0126 | No |
| 91 | SV2B | NA | SV2B Entrez, Source | synaptic vesicle glycoprotein 2B | 17293 | -0.115 | -0.0208 | No |
| 92 | PROP1 | NA | PROP1 Entrez, Source | prophet of Pit1, paired-like homeodomain transcription factor | 17749 | -0.137 | -0.0364 | No |
| 93 | FGGY | NA | 17769 | -0.138 | -0.0359 | No | ||
| 94 | CSRP3 | NA | CSRP3 Entrez, Source | cysteine and glycine-rich protein 3 (cardiac LIM protein) | 18076 | -0.157 | -0.0459 | No |
| 95 | ADAMTS20 | NA | ADAMTS20 Entrez, Source | ADAM metallopeptidase with thrombospondin type 1 motif, 20 | 18404 | -0.178 | -0.0564 | No |
| 96 | LATS1 | NA | LATS1 Entrez, Source | LATS, large tumor suppressor, homolog 1 (Drosophila) | 19021 | -0.229 | -0.0771 | No |
| 97 | MUC13 | NA | MUC13 Entrez, Source | mucin 13, cell surface associated | 19087 | -0.234 | -0.0776 | No |
| 98 | CLDN8 | NA | CLDN8 Entrez, Source | claudin 8 | 19181 | -0.242 | -0.0790 | No |
| 99 | TSHB | NA | TSHB Entrez, Source | thyroid stimulating hormone, beta | 19841 | -0.304 | -0.1007 | No |
| 100 | RAMP3 | NA | RAMP3 Entrez, Source | receptor (calcitonin) activity modifying protein 3 | 20165 | -0.338 | -0.1097 | No |
| 101 | RPL13P5 | NA | 20215 | -0.343 | -0.1086 | No | ||
| 102 | PVRL1 | NA | PVRL1 Entrez, Source | poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 20227 | -0.345 | -0.1062 | No |
| 103 | CCDC57 | NA | CCDC57 Entrez, Source | coiled-coil domain containing 57 | 20364 | -0.364 | -0.1081 | No |
| 104 | FGR | NA | FGR Entrez, Source | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | 20557 | -0.387 | -0.1119 | No |
| 105 | TEX15 | NA | TEX15 Entrez, Source | testis expressed sequence 15 | 20860 | -0.431 | -0.1194 | No |
| 106 | HPR | NA | HPR Entrez, Source | haptoglobin-related protein | 20926 | -0.441 | -0.1181 | No |
| 107 | CLSTN3 | NA | CLSTN3 Entrez, Source | calsyntenin 3 | 20943 | -0.444 | -0.1150 | No |
| 108 | MYOM2 | NA | MYOM2 Entrez, Source | myomesin (M-protein) 2, 165kDa | 21071 | -0.462 | -0.1158 | No |
| 109 | XYLB | NA | XYLB Entrez, Source | xylulokinase homolog (H. influenzae) | 21892 | -0.608 | -0.1409 | No |
| 110 | DTNB | NA | DTNB Entrez, Source | dystrobrevin, beta | 22228 | -0.671 | -0.1476 | No |
| 111 | EGFL7 | NA | EGFL7 Entrez, Source | EGF-like-domain, multiple 7 | 22291 | -0.686 | -0.1441 | No |
| 112 | CRH | NA | CRH Entrez, Source | corticotropin releasing hormone | 22400 | -0.708 | -0.1422 | No |
| 113 | HHLA1 | NA | HHLA1 Entrez, Source | HERV-H LTR-associating 1 | 22708 | -0.775 | -0.1470 | No |
| 114 | SIDT1 | NA | SIDT1 Entrez, Source | SID1 transmembrane family, member 1 | 22713 | -0.776 | -0.1407 | No |
| 115 | EGR4 | NA | EGR4 Entrez, Source | early growth response 4 | 22845 | -0.805 | -0.1388 | No |
| 116 | LOC440925 | NA | LOC440925 Entrez, Source | - | 23538 | -0.961 | -0.1562 | No |
| 117 | PI15 | NA | PI15 Entrez, Source | peptidase inhibitor 15 | 23943 | -1.075 | -0.1620 | No |
| 118 | KDM4D | NA | 23964 | -1.080 | -0.1538 | No | ||
| 119 | SLC8A1 | NA | SLC8A1 Entrez, Source | solute carrier family 8 (sodium/calcium exchanger), member 1 | 24111 | -1.124 | -0.1497 | No |
| 120 | TNNI3 | NA | TNNI3 Entrez, Source | troponin I type 3 (cardiac) | 24439 | -1.223 | -0.1515 | No |
| 121 | KIF25 | NA | KIF25 Entrez, Source | kinesin family member 25 | 24946 | -1.394 | -0.1585 | No |
| 122 | ALOX12B | NA | ALOX12B Entrez, Source | arachidonate 12-lipoxygenase, 12R type | 25048 | -1.437 | -0.1502 | No |
| 123 | IL18R1 | NA | IL18R1 Entrez, Source | interleukin 18 receptor 1 | 25136 | -1.478 | -0.1410 | No |
| 124 | RUNDC3B | NA | 25213 | -1.507 | -0.1312 | No | ||
| 125 | SPTBN2 | NA | SPTBN2 Entrez, Source | spectrin, beta, non-erythrocytic 2 | 25615 | -1.695 | -0.1318 | No |
| 126 | EDIL3 | NA | EDIL3 Entrez, Source | EGF-like repeats and discoidin I-like domains 3 | 25672 | -1.726 | -0.1194 | No |
| 127 | SMPDL3B | NA | SMPDL3B Entrez, Source | sphingomyelin phosphodiesterase, acid-like 3B | 25816 | -1.805 | -0.1096 | No |
| 128 | SLC9A7 | NA | SLC9A7 Entrez, Source | solute carrier family 9 (sodium/hydrogen exchanger), member 7 | 26189 | -2.050 | -0.1061 | No |
| 129 | TLR6 | NA | TLR6 Entrez, Source | toll-like receptor 6 | 26258 | -2.092 | -0.0912 | No |
| 130 | ASPHD1 | NA | ASPHD1 Entrez, Source | aspartate beta-hydroxylase domain containing 1 | 26501 | -2.302 | -0.0808 | No |
| 131 | EXOSC1 | NA | EXOSC1 Entrez, Source | exosome component 1 | 26751 | -2.561 | -0.0686 | No |
| 132 | C17orf59 | NA | C17ORF59 Entrez, Source | chromosome 17 open reading frame 59 | 26922 | -2.858 | -0.0509 | No |
| 133 | SLC30A3 | NA | SLC30A3 Entrez, Source | solute carrier family 30 (zinc transporter), member 3 | 27016 | -3.078 | -0.0286 | No |
| 134 | PLBD1 | NA | 27318 | -4.796 | 0.0004 | No |