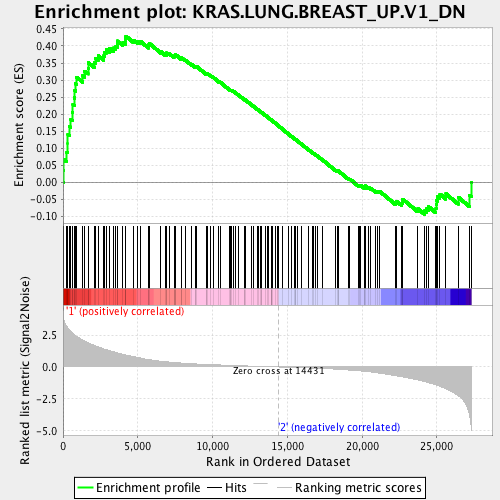
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | KRAS.LUNG.BREAST_UP.V1_DN |
| Enrichment Score (ES) | 0.42896348 |
| Normalized Enrichment Score (NES) | 1.6823387 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.007526414 |
| FWER p-Value | 0.039 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GADD45G | NA | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 25 | 3.828 | 0.0338 | Yes |
| 2 | IGFBP2 | NA | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 37 | 3.744 | 0.0674 | Yes |
| 3 | TRIM9 | NA | TRIM9 Entrez, Source | tripartite motif-containing 9 | 251 | 3.172 | 0.0883 | Yes |
| 4 | VAV3 | NA | VAV3 Entrez, Source | vav 3 oncogene | 319 | 3.047 | 0.1135 | Yes |
| 5 | IFI44L | NA | IFI44L Entrez, Source | interferon-induced protein 44-like | 322 | 3.045 | 0.1410 | Yes |
| 6 | METTL7A | NA | METTL7A Entrez, Source | methyltransferase like 7A | 433 | 2.889 | 0.1632 | Yes |
| 7 | CXCR7 | NA | CXCR7 Entrez, Source | chemokine (C-X-C motif) receptor 7 | 520 | 2.766 | 0.1851 | Yes |
| 8 | RBM17 | NA | RBM17 Entrez, Source | RNA binding motif protein 17 | 610 | 2.667 | 0.2061 | Yes |
| 9 | SOBP | NA | 657 | 2.617 | 0.2281 | Yes | ||
| 10 | PAQR6 | NA | PAQR6 Entrez, Source | progestin and adipoQ receptor family member VI | 750 | 2.521 | 0.2476 | Yes |
| 11 | MEGF8 | NA | MEGF8 Entrez, Source | multiple EGF-like-domains 8 | 786 | 2.489 | 0.2689 | Yes |
| 12 | RAG1 | NA | RAG1 Entrez, Source | recombination activating gene 1 | 842 | 2.440 | 0.2890 | Yes |
| 13 | PBX1 | NA | PBX1 Entrez, Source | pre-B-cell leukemia transcription factor 1 | 894 | 2.392 | 0.3088 | Yes |
| 14 | FRAS1 | NA | FRAS1 Entrez, Source | Fraser syndrome 1 | 1318 | 2.089 | 0.3122 | Yes |
| 15 | MXD3 | NA | MXD3 Entrez, Source | MAX dimerization protein 3 | 1436 | 2.013 | 0.3262 | Yes |
| 16 | DTX2 | NA | DTX2 Entrez, Source | deltex homolog 2 (Drosophila) | 1667 | 1.889 | 0.3349 | Yes |
| 17 | OLFML2A | NA | OLFML2A Entrez, Source | olfactomedin-like 2A | 1700 | 1.871 | 0.3507 | Yes |
| 18 | CD36 | NA | CD36 Entrez, Source | CD36 molecule (thrombospondin receptor) | 2108 | 1.674 | 0.3509 | Yes |
| 19 | EPB41L4A | NA | EPB41L4A Entrez, Source | erythrocyte membrane protein band 4.1 like 4A | 2155 | 1.653 | 0.3642 | Yes |
| 20 | NYNRIN | NA | 2340 | 1.580 | 0.3717 | Yes | ||
| 21 | EPHX2 | NA | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 2718 | 1.416 | 0.3707 | Yes |
| 22 | VAX2 | NA | VAX2 Entrez, Source | ventral anterior homeobox 2 | 2758 | 1.396 | 0.3820 | Yes |
| 23 | HCN2 | NA | HCN2 Entrez, Source | hyperpolarization activated cyclic nucleotide-gated potassium channel 2 | 2874 | 1.359 | 0.3901 | Yes |
| 24 | SOX11 | NA | SOX11 Entrez, Source | SRY (sex determining region Y)-box 11 | 3095 | 1.278 | 0.3936 | Yes |
| 25 | PIK3C2B | NA | PIK3C2B Entrez, Source | phosphoinositide-3-kinase, class 2, beta polypeptide | 3375 | 1.192 | 0.3941 | Yes |
| 26 | TTLL1 | NA | TTLL1 Entrez, Source | tubulin tyrosine ligase-like family, member 1 | 3532 | 1.144 | 0.3988 | Yes |
| 27 | PDE6B | NA | PDE6B Entrez, Source | phosphodiesterase 6B, cGMP-specific, rod, beta (congenital stationary night blindness 3, autosomal dominant) | 3624 | 1.108 | 0.4055 | Yes |
| 28 | AKR1B10 | NA | AKR1B10 Entrez, Source | aldo-keto reductase family 1, member B10 (aldose reductase) | 3645 | 1.100 | 0.4147 | Yes |
| 29 | ATP6V1B1 | NA | ATP6V1B1 Entrez, Source | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 (Renal tubular acidosis with deafness) | 4005 | 0.986 | 0.4105 | Yes |
| 30 | YPEL1 | NA | YPEL1 Entrez, Source | yippee-like 1 (Drosophila) | 4152 | 0.945 | 0.4137 | Yes |
| 31 | RTP4 | NA | RTP4 Entrez, Source | receptor transporter protein 4 | 4159 | 0.942 | 0.4220 | Yes |
| 32 | MAP2K6 | NA | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 4200 | 0.932 | 0.4290 | Yes |
| 33 | ALDH5A1 | NA | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 4715 | 0.806 | 0.4174 | No |
| 34 | CXCL14 | NA | CXCL14 Entrez, Source | chemokine (C-X-C motif) ligand 14 | 4987 | 0.746 | 0.4142 | No |
| 35 | AKAP6 | NA | AKAP6 Entrez, Source | A kinase (PRKA) anchor protein 6 | 5156 | 0.707 | 0.4144 | No |
| 36 | INPP5D | NA | INPP5D Entrez, Source | inositol polyphosphate-5-phosphatase, 145kDa | 5714 | 0.576 | 0.3991 | No |
| 37 | ADD2 | NA | ADD2 Entrez, Source | adducin 2 (beta) | 5740 | 0.572 | 0.4034 | No |
| 38 | ATHL1 | NA | ATHL1 Entrez, Source | ATH1, acid trehalase-like 1 (yeast) | 5782 | 0.563 | 0.4070 | No |
| 39 | NSUN6 | NA | NSUN6 Entrez, Source | NOL1/NOP2/Sun domain family, member 6 | 6548 | 0.447 | 0.3829 | No |
| 40 | IVL | NA | IVL Entrez, Source | involucrin | 6836 | 0.415 | 0.3762 | No |
| 41 | CDH12 | NA | CDH12 Entrez, Source | cadherin 12, type 2 (N-cadherin 2) | 6932 | 0.404 | 0.3763 | No |
| 42 | MTHFR | NA | MTHFR Entrez, Source | 5,10-methylenetetrahydrofolate reductase (NADPH) | 6944 | 0.403 | 0.3796 | No |
| 43 | HTR1B | NA | HTR1B Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 1B | 7098 | 0.386 | 0.3775 | No |
| 44 | PROC | NA | PROC Entrez, Source | protein C (inactivator of coagulation factors Va and VIIIa) | 7423 | 0.352 | 0.3687 | No |
| 45 | COL2A1 | NA | COL2A1 Entrez, Source | collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 7453 | 0.349 | 0.3708 | No |
| 46 | C6orf10 | NA | C6ORF10 Entrez, Source | chromosome 6 open reading frame 10 | 7528 | 0.342 | 0.3712 | No |
| 47 | GPR77 | NA | GPR77 Entrez, Source | G protein-coupled receptor 77 | 7536 | 0.341 | 0.3740 | No |
| 48 | GLS2 | NA | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 7913 | 0.308 | 0.3630 | No |
| 49 | ART1 | NA | ART1 Entrez, Source | ADP-ribosyltransferase 1 | 7921 | 0.308 | 0.3655 | No |
| 50 | ERAP2 | NA | 8182 | 0.286 | 0.3586 | No | ||
| 51 | CCDC132 | NA | CCDC132 Entrez, Source | coiled-coil domain containing 132 | 8607 | 0.256 | 0.3453 | No |
| 52 | TAS2R4 | NA | TAS2R4 Entrez, Source | taste receptor, type 2, member 4 | 8862 | 0.238 | 0.3381 | No |
| 53 | NCR3 | NA | NCR3 Entrez, Source | natural cytotoxicity triggering receptor 3 | 8890 | 0.237 | 0.3393 | No |
| 54 | TRIM48 | NA | TRIM48 Entrez, Source | tripartite motif-containing 48 | 8914 | 0.235 | 0.3406 | No |
| 55 | COL5A2 | NA | COL5A2 Entrez, Source | collagen, type V, alpha 2 | 9563 | 0.200 | 0.3186 | No |
| 56 | MMP28 | NA | MMP28 Entrez, Source | matrix metallopeptidase 28 | 9616 | 0.197 | 0.3184 | No |
| 57 | KRT13 | NA | KRT13 Entrez, Source | keratin 13 | 9639 | 0.195 | 0.3194 | No |
| 58 | LGALS7 | NA | LGALS7 Entrez, Source | lectin, galactoside-binding, soluble, 7 (galectin 7) | 9838 | 0.186 | 0.3138 | No |
| 59 | HIST1H3A | NA | HIST1H3A Entrez, Source | histone cluster 1, H3a | 10046 | 0.177 | 0.3078 | No |
| 60 | PDE3A | NA | PDE3A Entrez, Source | phosphodiesterase 3A, cGMP-inhibited | 10425 | 0.159 | 0.2953 | No |
| 61 | CACNA1I | NA | CACNA1I Entrez, Source | calcium channel, voltage-dependent, alpha 1I subunit | 10499 | 0.156 | 0.2941 | No |
| 62 | NOX5 | NA | NOX5 Entrez, Source | NADPH oxidase, EF-hand calcium binding domain 5 | 11143 | 0.126 | 0.2716 | No |
| 63 | FGFR3 | NA | FGFR3 Entrez, Source | fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) | 11229 | 0.122 | 0.2695 | No |
| 64 | IFNA16 | NA | IFNA16 Entrez, Source | interferon, alpha 16 | 11230 | 0.122 | 0.2706 | No |
| 65 | SLC6A9 | NA | SLC6A9 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 11257 | 0.120 | 0.2708 | No |
| 66 | KRT16 | NA | KRT16 Entrez, Source | keratin 16 (focal non-epidermolytic palmoplantar keratoderma) | 11408 | 0.113 | 0.2663 | No |
| 67 | CYP21A2 | NA | CYP21A2 Entrez, Source | cytochrome P450, family 21, subfamily A, polypeptide 2 | 11564 | 0.108 | 0.2616 | No |
| 68 | FLT1 | NA | FLT1 Entrez, Source | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 11746 | 0.100 | 0.2558 | No |
| 69 | CD248 | NA | CD248 Entrez, Source | CD248 molecule, endosialin | 12102 | 0.087 | 0.2435 | No |
| 70 | DEFB1 | NA | DEFB1 Entrez, Source | defensin, beta 1 | 12165 | 0.084 | 0.2420 | No |
| 71 | GABRA4 | NA | GABRA4 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 4 | 12200 | 0.083 | 0.2415 | No |
| 72 | LGALS9 | NA | LGALS9 Entrez, Source | lectin, galactoside-binding, soluble, 9 (galectin 9) | 12602 | 0.067 | 0.2274 | No |
| 73 | CCDC121 | NA | CCDC121 Entrez, Source | coiled-coil domain containing 121 | 12714 | 0.063 | 0.2239 | No |
| 74 | CALML5 | NA | CALML5 Entrez, Source | calmodulin-like 5 | 13006 | 0.053 | 0.2137 | No |
| 75 | MYL9 | NA | MYL9 Entrez, Source | myosin, light chain 9, regulatory | 13092 | 0.049 | 0.2110 | No |
| 76 | APOB | NA | APOB Entrez, Source | apolipoprotein B (including Ag(x) antigen) | 13212 | 0.045 | 0.2070 | No |
| 77 | ANKRD1 | NA | ANKRD1 Entrez, Source | ankyrin repeat domain 1 (cardiac muscle) | 13268 | 0.042 | 0.2054 | No |
| 78 | TBR1 | NA | TBR1 Entrez, Source | T-box, brain, 1 | 13304 | 0.041 | 0.2045 | No |
| 79 | SCN7A | NA | SCN7A Entrez, Source | sodium channel, voltage-gated, type VII, alpha | 13567 | 0.030 | 0.1951 | No |
| 80 | SLC12A3 | NA | SLC12A3 Entrez, Source | solute carrier family 12 (sodium/chloride transporters), member 3 | 13704 | 0.026 | 0.1903 | No |
| 81 | CDH16 | NA | CDH16 Entrez, Source | cadherin 16, KSP-cadherin | 13744 | 0.024 | 0.1891 | No |
| 82 | DPT | NA | DPT Entrez, Source | dermatopontin | 13914 | 0.018 | 0.1831 | No |
| 83 | KRT4 | NA | KRT4 Entrez, Source | keratin 4 | 13946 | 0.017 | 0.1821 | No |
| 84 | MAST3 | NA | MAST3 Entrez, Source | microtubule associated serine/threonine kinase 3 | 13974 | 0.016 | 0.1812 | No |
| 85 | MXRA8 | NA | MXRA8 Entrez, Source | matrix-remodelling associated 8 | 14008 | 0.015 | 0.1802 | No |
| 86 | TM4SF4 | NA | TM4SF4 Entrez, Source | transmembrane 4 L six family member 4 | 14180 | 0.009 | 0.1739 | No |
| 87 | UPK3B | NA | UPK3B Entrez, Source | uroplakin 3B | 14339 | 0.003 | 0.1682 | No |
| 88 | NOS1 | NA | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 14403 | 0.001 | 0.1659 | No |
| 89 | GDNF | NA | GDNF Entrez, Source | glial cell derived neurotrophic factor | 14707 | -0.010 | 0.1548 | No |
| 90 | KCNMB1 | NA | KCNMB1 Entrez, Source | potassium large conductance calcium-activated channel, subfamily M, beta member 1 | 15112 | -0.026 | 0.1402 | No |
| 91 | NTF3 | NA | NTF3 Entrez, Source | neurotrophin 3 | 15279 | -0.032 | 0.1344 | No |
| 92 | TNFSF10 | NA | TNFSF10 Entrez, Source | tumor necrosis factor (ligand) superfamily, member 10 | 15485 | -0.040 | 0.1272 | No |
| 93 | SERPINA10 | NA | SERPINA10 Entrez, Source | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 | 15534 | -0.042 | 0.1258 | No |
| 94 | THNSL2 | NA | 15564 | -0.043 | 0.1251 | No | ||
| 95 | KRT38 | NA | KRT38 Entrez, Source | keratin 38 | 15700 | -0.048 | 0.1206 | No |
| 96 | PCDHB1 | NA | PCDHB1 Entrez, Source | protocadherin beta 1 | 15980 | -0.058 | 0.1109 | No |
| 97 | P2RX1 | NA | P2RX1 Entrez, Source | purinergic receptor P2X, ligand-gated ion channel, 1 | 16433 | -0.079 | 0.0950 | No |
| 98 | BDKRB1 | NA | BDKRB1 Entrez, Source | bradykinin receptor B1 | 16675 | -0.088 | 0.0869 | No |
| 99 | RARRES2 | NA | RARRES2 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 2 | 16728 | -0.090 | 0.0858 | No |
| 100 | FMO6P | NA | 16855 | -0.096 | 0.0820 | No | ||
| 101 | IL5RA | NA | IL5RA Entrez, Source | interleukin 5 receptor, alpha | 17007 | -0.102 | 0.0774 | No |
| 102 | PVALB | NA | PVALB Entrez, Source | parvalbumin | 17366 | -0.118 | 0.0653 | No |
| 103 | S100A7 | NA | S100A7 Entrez, Source | S100 calcium binding protein A7 | 18221 | -0.167 | 0.0354 | No |
| 104 | SERPINB13 | NA | SERPINB13 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 13 | 18338 | -0.174 | 0.0327 | No |
| 105 | RYR1 | NA | RYR1 Entrez, Source | ryanodine receptor 1 (skeletal) | 18403 | -0.178 | 0.0320 | No |
| 106 | ADAMTS20 | NA | ADAMTS20 Entrez, Source | ADAM metallopeptidase with thrombospondin type 1 motif, 20 | 18404 | -0.178 | 0.0336 | No |
| 107 | HTR1E | NA | HTR1E Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 1E | 19108 | -0.236 | 0.0099 | No |
| 108 | CLDN8 | NA | CLDN8 Entrez, Source | claudin 8 | 19181 | -0.242 | 0.0095 | No |
| 109 | LY6D | NA | LY6D Entrez, Source | lymphocyte antigen 6 complex, locus D | 19787 | -0.298 | -0.0101 | No |
| 110 | TSHB | NA | TSHB Entrez, Source | thyroid stimulating hormone, beta | 19841 | -0.304 | -0.0093 | No |
| 111 | CLCA2 | NA | CLCA2 Entrez, Source | chloride channel, calcium activated, family member 2 | 19909 | -0.310 | -0.0089 | No |
| 112 | RAMP3 | NA | RAMP3 Entrez, Source | receptor (calcitonin) activity modifying protein 3 | 20165 | -0.338 | -0.0152 | No |
| 113 | CALML3 | NA | CALML3 Entrez, Source | calmodulin-like 3 | 20178 | -0.340 | -0.0126 | No |
| 114 | RPL13P5 | NA | 20215 | -0.343 | -0.0108 | No | ||
| 115 | SLC3A1 | NA | SLC3A1 Entrez, Source | 1 | 20428 | -0.371 | -0.0152 | No |
| 116 | AMBN | NA | AMBN Entrez, Source | ameloblastin (enamel matrix protein) | 20590 | -0.391 | -0.0176 | No |
| 117 | SCARF1 | NA | SCARF1 Entrez, Source | scavenger receptor class F, member 1 | 20922 | -0.440 | -0.0258 | No |
| 118 | GLYAT | NA | GLYAT Entrez, Source | glycine-N-acyltransferase | 21048 | -0.460 | -0.0262 | No |
| 119 | CD6 | NA | CD6 Entrez, Source | CD6 molecule | 21181 | -0.478 | -0.0267 | No |
| 120 | DTNB | NA | DTNB Entrez, Source | dystrobrevin, beta | 22228 | -0.671 | -0.0591 | No |
| 121 | GAMT | NA | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 22321 | -0.691 | -0.0562 | No |
| 122 | TGM1 | NA | TGM1 Entrez, Source | transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) | 22653 | -0.766 | -0.0614 | No |
| 123 | HHLA1 | NA | HHLA1 Entrez, Source | HERV-H LTR-associating 1 | 22708 | -0.775 | -0.0564 | No |
| 124 | SIDT1 | NA | SIDT1 Entrez, Source | SID1 transmembrane family, member 1 | 22713 | -0.776 | -0.0495 | No |
| 125 | PARP3 | NA | PARP3 Entrez, Source | poly (ADP-ribose) polymerase family, member 3 | 23742 | -1.017 | -0.0781 | No |
| 126 | COBL | NA | COBL Entrez, Source | cordon-bleu homolog (mouse) | 24195 | -1.147 | -0.0843 | No |
| 127 | HRK | NA | HRK Entrez, Source | harakiri, BCL2 interacting protein (contains only BH3 domain) | 24327 | -1.188 | -0.0783 | No |
| 128 | TNNI3 | NA | TNNI3 Entrez, Source | troponin I type 3 (cardiac) | 24439 | -1.223 | -0.0713 | No |
| 129 | KIF25 | NA | KIF25 Entrez, Source | kinesin family member 25 | 24946 | -1.394 | -0.0773 | No |
| 130 | CRABP2 | NA | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 24956 | -1.400 | -0.0649 | No |
| 131 | TRPM2 | NA | TRPM2 Entrez, Source | transient receptor potential cation channel, subfamily M, member 2 | 24983 | -1.411 | -0.0531 | No |
| 132 | ALOX12B | NA | ALOX12B Entrez, Source | arachidonate 12-lipoxygenase, 12R type | 25048 | -1.437 | -0.0424 | No |
| 133 | RUNDC3B | NA | 25213 | -1.507 | -0.0347 | No | ||
| 134 | SPTBN2 | NA | SPTBN2 Entrez, Source | spectrin, beta, non-erythrocytic 2 | 25615 | -1.695 | -0.0341 | No |
| 135 | PTPRU | NA | PTPRU Entrez, Source | protein tyrosine phosphatase, receptor type, U | 26431 | -2.240 | -0.0438 | No |
| 136 | EMR1 | NA | EMR1 Entrez, Source | egf-like module containing, mucin-like, hormone receptor-like 1 | 27173 | -3.659 | -0.0378 | No |
| 137 | PLBD1 | NA | 27318 | -4.796 | 0.0004 | No |