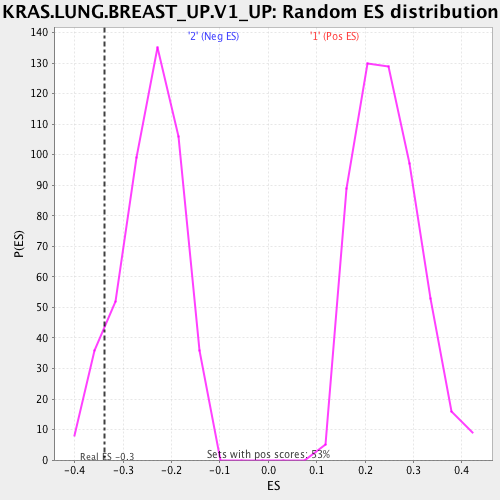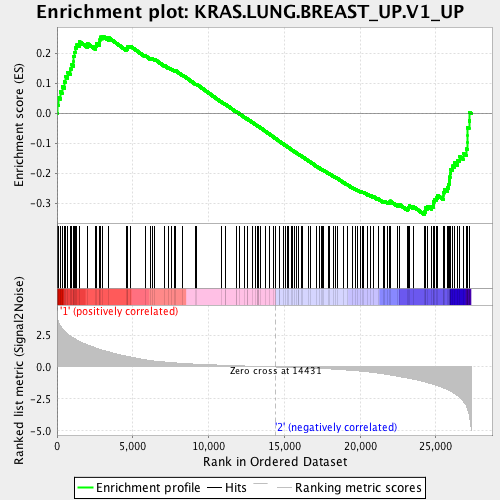
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | KRAS.LUNG.BREAST_UP.V1_UP |
| Enrichment Score (ES) | -0.337983 |
| Normalized Enrichment Score (NES) | -1.3895284 |
| Nominal p-value | 0.08898305 |
| FDR q-value | 0.16297361 |
| FWER p-Value | 0.55 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SPRY2 | NA | SPRY2 Entrez, Source | sprouty homolog 2 (Drosophila) | 9 | 3.989 | 0.0292 | No |
| 2 | DNAH9 | NA | DNAH9 Entrez, Source | dynein, axonemal, heavy chain 9 | 94 | 3.503 | 0.0521 | No |
| 3 | IGFBP3 | NA | IGFBP3 Entrez, Source | insulin-like growth factor binding protein 3 | 206 | 3.262 | 0.0722 | No |
| 4 | GULP1 | NA | GULP1 Entrez, Source | GULP, engulfment adaptor PTB domain containing 1 | 330 | 3.029 | 0.0901 | No |
| 5 | ID2 | NA | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 481 | 2.829 | 0.1055 | No |
| 6 | SNAP25 | NA | SNAP25 Entrez, Source | synaptosomal-associated protein, 25kDa | 546 | 2.722 | 0.1233 | No |
| 7 | KCNC1 | NA | KCNC1 Entrez, Source | potassium voltage-gated channel, Shaw-related subfamily, member 1 | 698 | 2.579 | 0.1369 | No |
| 8 | ANGPTL4 | NA | ANGPTL4 Entrez, Source | angiopoietin-like 4 | 866 | 2.417 | 0.1486 | No |
| 9 | ST3GAL6 | NA | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 923 | 2.370 | 0.1641 | No |
| 10 | MGLL | NA | MGLL Entrez, Source | monoglyceride lipase | 1086 | 2.251 | 0.1749 | No |
| 11 | KIF5C | NA | KIF5C Entrez, Source | kinesin family member 5C | 1092 | 2.248 | 0.1913 | No |
| 12 | C11orf41 | NA | C11ORF41 Entrez, Source | chromosome 11 open reading frame 41 | 1164 | 2.200 | 0.2050 | No |
| 13 | TAGLN3 | NA | TAGLN3 Entrez, Source | transgelin 3 | 1188 | 2.185 | 0.2204 | No |
| 14 | HOXD11 | NA | HOXD11 Entrez, Source | homeobox D11 | 1307 | 2.095 | 0.2315 | No |
| 15 | HAS2 | NA | HAS2 Entrez, Source | hyaluronan synthase 2 | 1487 | 1.986 | 0.2397 | No |
| 16 | DOCK4 | NA | DOCK4 Entrez, Source | dedicator of cytokinesis 4 | 1989 | 1.731 | 0.2341 | No |
| 17 | AKT2 | NA | AKT2 Entrez, Source | v-akt murine thymoma viral oncogene homolog 2 | 2555 | 1.479 | 0.2242 | No |
| 18 | ADAM19 | NA | ADAM19 Entrez, Source | ADAM metallopeptidase domain 19 (meltrin beta) | 2593 | 1.464 | 0.2337 | No |
| 19 | NDOR1 | NA | NDOR1 Entrez, Source | NADPH dependent diflavin oxidoreductase 1 | 2809 | 1.383 | 0.2361 | No |
| 20 | FAM174B | NA | 2825 | 1.377 | 0.2457 | No | ||
| 21 | RNASE1 | NA | RNASE1 Entrez, Source | ribonuclease, RNase A family, 1 (pancreatic) | 2834 | 1.373 | 0.2556 | No |
| 22 | SEMA3B | NA | SEMA3B Entrez, Source | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B | 3019 | 1.304 | 0.2585 | No |
| 23 | C1orf114 | NA | C1ORF114 Entrez, Source | chromosome 1 open reading frame 114 | 3399 | 1.184 | 0.2533 | No |
| 24 | IL13RA2 | NA | IL13RA2 Entrez, Source | interleukin 13 receptor, alpha 2 | 4557 | 0.849 | 0.2171 | No |
| 25 | DKFZp566H0824 | NA | DKFZP566H0824 Entrez, Source | - | 4637 | 0.830 | 0.2203 | No |
| 26 | HBEGF | NA | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 4667 | 0.821 | 0.2253 | No |
| 27 | ZNF639 | NA | ZNF639 Entrez, Source | zinc finger protein 639 | 4845 | 0.777 | 0.2246 | No |
| 28 | NRL | NA | NRL Entrez, Source | neural retina leucine zipper | 5810 | 0.559 | 0.1933 | No |
| 29 | A2M | NA | A2M Entrez, Source | alpha-2-macroglobulin | 6145 | 0.506 | 0.1847 | No |
| 30 | TNFSF15 | NA | TNFSF15 Entrez, Source | tumor necrosis factor (ligand) superfamily, member 15 | 6281 | 0.485 | 0.1833 | No |
| 31 | KIF5A | NA | KIF5A Entrez, Source | kinesin family member 5A | 6447 | 0.458 | 0.1807 | No |
| 32 | CRCT1 | NA | 7092 | 0.387 | 0.1599 | No | ||
| 33 | CD37 | NA | CD37 Entrez, Source | CD37 molecule | 7375 | 0.358 | 0.1521 | No |
| 34 | LDB3 | NA | LDB3 Entrez, Source | LIM domain binding 3 | 7560 | 0.339 | 0.1479 | No |
| 35 | HPN | NA | HPN Entrez, Source | hepsin (transmembrane protease, serine 1) | 7770 | 0.321 | 0.1426 | No |
| 36 | POLM | NA | POLM Entrez, Source | polymerase (DNA directed), mu | 7835 | 0.315 | 0.1425 | No |
| 37 | RBP4 | NA | RBP4 Entrez, Source | retinol binding protein 4, plasma | 8284 | 0.278 | 0.1281 | No |
| 38 | PTGS2 | NA | PTGS2 Entrez, Source | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 9159 | 0.221 | 0.0976 | No |
| 39 | TTC22 | NA | TTC22 Entrez, Source | tetratricopeptide repeat domain 22 | 9223 | 0.218 | 0.0969 | No |
| 40 | FNDC8 | NA | FNDC8 Entrez, Source | fibronectin type III domain containing 8 | 10863 | 0.139 | 0.0377 | No |
| 41 | GPR4 | NA | GPR4 Entrez, Source | G protein-coupled receptor 4 | 11084 | 0.128 | 0.0305 | No |
| 42 | OR2B6 | NA | OR2B6 Entrez, Source | olfactory receptor, family 2, subfamily B, member 6 | 11131 | 0.126 | 0.0298 | No |
| 43 | INHBA | NA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 11814 | 0.097 | 0.0054 | No |
| 44 | NPPB | NA | NPPB Entrez, Source | natriuretic peptide precursor B | 11836 | 0.096 | 0.0053 | No |
| 45 | AMELY | NA | AMELY Entrez, Source | amelogenin, Y-linked | 12059 | 0.088 | -0.0022 | No |
| 46 | CCL20 | NA | CCL20 Entrez, Source | chemokine (C-C motif) ligand 20 | 12374 | 0.076 | -0.0132 | No |
| 47 | CD247 | NA | CD247 Entrez, Source | CD247 molecule | 12530 | 0.070 | -0.0183 | No |
| 48 | TFDP3 | NA | TFDP3 Entrez, Source | transcription factor Dp family, member 3 | 12565 | 0.069 | -0.0191 | No |
| 49 | LILRA6 | NA | LILRA6 Entrez, Source | leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 | 12898 | 0.057 | -0.0309 | No |
| 50 | CEACAM3 | NA | CEACAM3 Entrez, Source | carcinoembryonic antigen-related cell adhesion molecule 3 | 13095 | 0.049 | -0.0377 | No |
| 51 | OSGIN1 | NA | OSGIN1 Entrez, Source | oxidative stress induced growth inhibitor 1 | 13198 | 0.045 | -0.0411 | No |
| 52 | MYH2 | NA | MYH2 Entrez, Source | myosin, heavy chain 2, skeletal muscle, adult | 13245 | 0.043 | -0.0425 | No |
| 53 | RBMXL2 | NA | 13279 | 0.042 | -0.0434 | No | ||
| 54 | LAT2 | NA | LAT2 Entrez, Source | linker for activation of T cells family, member 2 | 13288 | 0.042 | -0.0434 | No |
| 55 | CLSTN2 | NA | CLSTN2 Entrez, Source | calsyntenin 2 | 13398 | 0.037 | -0.0471 | No |
| 56 | PPBP | NA | PPBP Entrez, Source | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | 13740 | 0.024 | -0.0595 | No |
| 57 | CMKLR1 | NA | CMKLR1 Entrez, Source | chemokine-like receptor 1 | 14027 | 0.014 | -0.0699 | No |
| 58 | RELN | NA | RELN Entrez, Source | reelin | 14279 | 0.005 | -0.0791 | No |
| 59 | HSD17B2 | NA | HSD17B2 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 2 | 14388 | 0.002 | -0.0830 | No |
| 60 | RETN | NA | RETN Entrez, Source | resistin | 14399 | 0.001 | -0.0834 | No |
| 61 | CDSN | NA | CDSN Entrez, Source | corneodesmosin | 14666 | -0.008 | -0.0931 | No |
| 62 | SHOX | NA | SHOX Entrez, Source | short stature homeobox | 14673 | -0.008 | -0.0933 | No |
| 63 | PRSS3 | NA | PRSS3 Entrez, Source | protease, serine, 3 (mesotrypsin) | 14912 | -0.017 | -0.1019 | No |
| 64 | NR1H4 | NA | NR1H4 Entrez, Source | nuclear receptor subfamily 1, group H, member 4 | 15069 | -0.024 | -0.1075 | No |
| 65 | ESM1 | NA | ESM1 Entrez, Source | endothelial cell-specific molecule 1 | 15233 | -0.030 | -0.1132 | No |
| 66 | RMST | NA | RMST Entrez, Source | rhabdomyosarcoma 2 associated transcript (non-coding RNA) | 15239 | -0.031 | -0.1132 | No |
| 67 | CCR6 | NA | CCR6 Entrez, Source | chemokine (C-C motif) receptor 6 | 15455 | -0.039 | -0.1208 | No |
| 68 | FCRL2 | NA | FCRL2 Entrez, Source | Fc receptor-like 2 | 15522 | -0.042 | -0.1229 | No |
| 69 | IL1R2 | NA | IL1R2 Entrez, Source | interleukin 1 receptor, type II | 15692 | -0.048 | -0.1288 | No |
| 70 | SLC25A31 | NA | SLC25A31 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 | 15810 | -0.052 | -0.1327 | No |
| 71 | IL8 | NA | IL8 Entrez, Source | interleukin 8 | 15907 | -0.056 | -0.1358 | No |
| 72 | GRP | NA | GRP Entrez, Source | gastrin-releasing peptide | 15949 | -0.057 | -0.1369 | No |
| 73 | KCNK15 | NA | KCNK15 Entrez, Source | potassium channel, subfamily K, member 15 | 16137 | -0.066 | -0.1433 | No |
| 74 | CSF2 | NA | CSF2 Entrez, Source | colony stimulating factor 2 (granulocyte-macrophage) | 16165 | -0.067 | -0.1438 | No |
| 75 | IFNA8 | NA | IFNA8 Entrez, Source | interferon, alpha 8 | 16554 | -0.083 | -0.1574 | No |
| 76 | DHRS9 | NA | DHRS9 Entrez, Source | dehydrogenase/reductase (SDR family) member 9 | 16733 | -0.090 | -0.1633 | No |
| 77 | TAAR5 | NA | TAAR5 Entrez, Source | trace amine associated receptor 5 | 17118 | -0.106 | -0.1766 | No |
| 78 | C9orf38 | NA | C9ORF38 Entrez, Source | chromosome 9 open reading frame 38 | 17286 | -0.114 | -0.1819 | No |
| 79 | CLPS | NA | CLPS Entrez, Source | colipase, pancreatic | 17432 | -0.121 | -0.1864 | No |
| 80 | KCNC2 | NA | KCNC2 Entrez, Source | potassium voltage-gated channel, Shaw-related subfamily, member 2 | 17542 | -0.126 | -0.1895 | No |
| 81 | CALB1 | NA | CALB1 Entrez, Source | calbindin 1, 28kDa | 17578 | -0.128 | -0.1898 | No |
| 82 | HSD11B1 | NA | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 17896 | -0.147 | -0.2004 | No |
| 83 | SPINK1 | NA | SPINK1 Entrez, Source | serine peptidase inhibitor, Kazal type 1 | 18001 | -0.153 | -0.2031 | No |
| 84 | PRL | NA | PRL Entrez, Source | prolactin | 18243 | -0.168 | -0.2107 | No |
| 85 | CD1A | NA | CD1A Entrez, Source | CD1a molecule | 18355 | -0.175 | -0.2135 | No |
| 86 | RPS4Y1 | NA | RPS4Y1 Entrez, Source | ribosomal protein S4, Y-linked 1 | 18492 | -0.185 | -0.2171 | No |
| 87 | CHRNA2 | NA | CHRNA2 Entrez, Source | cholinergic receptor, nicotinic, alpha 2 (neuronal) | 18918 | -0.219 | -0.2311 | No |
| 88 | GBA3 | NA | GBA3 Entrez, Source | glucosidase, beta, acid 3 (cytosolic) | 19138 | -0.238 | -0.2374 | No |
| 89 | IL1RL1 | NA | IL1RL1 Entrez, Source | interleukin 1 receptor-like 1 | 19520 | -0.272 | -0.2494 | No |
| 90 | SMPX | NA | SMPX Entrez, Source | small muscle protein, X-linked | 19685 | -0.288 | -0.2533 | No |
| 91 | NTM | NA | 19833 | -0.303 | -0.2564 | No | ||
| 92 | ZP4 | NA | ZP4 Entrez, Source | zona pellucida glycoprotein 4 | 20019 | -0.322 | -0.2609 | No |
| 93 | ITGA2 | NA | ITGA2 Entrez, Source | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 20150 | -0.337 | -0.2631 | No |
| 94 | PRDM1 | NA | PRDM1 Entrez, Source | PR domain containing 1, with ZNF domain | 20217 | -0.343 | -0.2630 | No |
| 95 | MMP10 | NA | MMP10 Entrez, Source | matrix metallopeptidase 10 (stromelysin 2) | 20507 | -0.381 | -0.2708 | No |
| 96 | ATG10 | NA | ATG10 Entrez, Source | ATG10 autophagy related 10 homolog (S. cerevisiae) | 20685 | -0.404 | -0.2743 | No |
| 97 | ADAM8 | NA | ADAM8 Entrez, Source | ADAM metallopeptidase domain 8 | 20906 | -0.438 | -0.2792 | No |
| 98 | PDCD1LG2 | NA | PDCD1LG2 Entrez, Source | programmed cell death 1 ligand 2 | 21180 | -0.478 | -0.2857 | No |
| 99 | SH2D2A | NA | SH2D2A Entrez, Source | SH2 domain protein 2A | 21557 | -0.545 | -0.2955 | No |
| 100 | OSBP2 | NA | OSBP2 Entrez, Source | oxysterol binding protein 2 | 21630 | -0.556 | -0.2940 | No |
| 101 | CTNNA2 | NA | CTNNA2 Entrez, Source | catenin (cadherin-associated protein), alpha 2 | 21819 | -0.595 | -0.2965 | No |
| 102 | MKRN3 | NA | MKRN3 Entrez, Source | makorin, ring finger protein, 3 | 21929 | -0.614 | -0.2960 | No |
| 103 | TRAF1 | NA | TRAF1 Entrez, Source | TNF receptor-associated factor 1 | 21969 | -0.624 | -0.2928 | No |
| 104 | NELL2 | NA | NELL2 Entrez, Source | NEL-like 2 (chicken) | 22466 | -0.721 | -0.3057 | No |
| 105 | ITPKA | NA | ITPKA Entrez, Source | inositol 1,4,5-trisphosphate 3-kinase A | 22597 | -0.753 | -0.3049 | No |
| 106 | SCG5 | NA | SCG5 Entrez, Source | secretogranin V (7B2 protein) | 23141 | -0.867 | -0.3184 | No |
| 107 | KCNJ14 | NA | KCNJ14 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 14 | 23202 | -0.880 | -0.3141 | No |
| 108 | TNFAIP3 | NA | TNFAIP3 Entrez, Source | tumor necrosis factor, alpha-induced protein 3 | 23231 | -0.887 | -0.3086 | No |
| 109 | TSPAN1 | NA | TSPAN1 Entrez, Source | tetraspanin 1 | 23502 | -0.951 | -0.3115 | No |
| 110 | ETS1 | NA | ETS1 Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 24224 | -1.157 | -0.3294 | Yes |
| 111 | KRT34 | NA | KRT34 Entrez, Source | keratin 34 | 24309 | -1.182 | -0.3237 | Yes |
| 112 | STC1 | NA | STC1 Entrez, Source | stanniocalcin 1 | 24330 | -1.189 | -0.3157 | Yes |
| 113 | CSF3 | NA | CSF3 Entrez, Source | colony stimulating factor 3 (granulocyte) | 24446 | -1.225 | -0.3108 | Yes |
| 114 | SNAP91 | NA | SNAP91 Entrez, Source | synaptosomal-associated protein, 91kDa homolog (mouse) | 24696 | -1.307 | -0.3103 | Yes |
| 115 | CXCL3 | NA | CXCL3 Entrez, Source | chemokine (C-X-C motif) ligand 3 | 24820 | -1.350 | -0.3048 | Yes |
| 116 | GABRA3 | NA | GABRA3 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | 24833 | -1.355 | -0.2952 | Yes |
| 117 | CXCL2 | NA | CXCL2 Entrez, Source | chemokine (C-X-C motif) ligand 2 | 24873 | -1.368 | -0.2865 | Yes |
| 118 | BMP6 | NA | BMP6 Entrez, Source | bone morphogenetic protein 6 | 25012 | -1.421 | -0.2811 | Yes |
| 119 | PTPRR | NA | PTPRR Entrez, Source | protein tyrosine phosphatase, receptor type, R | 25101 | -1.465 | -0.2735 | Yes |
| 120 | SLCO5A1 | NA | SLCO5A1 Entrez, Source | solute carrier organic anion transporter family, member 5A1 | 25484 | -1.634 | -0.2754 | Yes |
| 121 | TBX3 | NA | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 25487 | -1.638 | -0.2634 | Yes |
| 122 | GLRX | NA | GLRX Entrez, Source | glutaredoxin (thioltransferase) | 25564 | -1.671 | -0.2538 | Yes |
| 123 | BIRC3 | NA | BIRC3 Entrez, Source | baculoviral IAP repeat-containing 3 | 25763 | -1.775 | -0.2479 | Yes |
| 124 | CXCL1 | NA | CXCL1 Entrez, Source | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 25823 | -1.806 | -0.2367 | Yes |
| 125 | GYPC | NA | GYPC Entrez, Source | glycophorin C (Gerbich blood group) | 25867 | -1.839 | -0.2247 | Yes |
| 126 | IL23A | NA | IL23A Entrez, Source | interleukin 23, alpha subunit p19 | 25913 | -1.862 | -0.2125 | Yes |
| 127 | SLCO4A1 | NA | SLCO4A1 Entrez, Source | solute carrier organic anion transporter family, member 4A1 | 25930 | -1.874 | -0.1992 | Yes |
| 128 | MMP9 | NA | MMP9 Entrez, Source | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 25971 | -1.898 | -0.1866 | Yes |
| 129 | LIF | NA | LIF Entrez, Source | leukemia inhibitory factor (cholinergic differentiation factor) | 26070 | -1.959 | -0.1757 | Yes |
| 130 | GFI1 | NA | GFI1 Entrez, Source | growth factor independent 1 | 26200 | -2.059 | -0.1652 | Yes |
| 131 | ADAMTS6 | NA | ADAMTS6 Entrez, Source | ADAM metallopeptidase with thrombospondin type 1 motif, 6 | 26418 | -2.226 | -0.1567 | Yes |
| 132 | TERT | NA | TERT Entrez, Source | telomerase reverse transcriptase | 26573 | -2.357 | -0.1449 | Yes |
| 133 | G0S2 | NA | G0S2 Entrez, Source | G0/G1switch 2 | 26789 | -2.628 | -0.1333 | Yes |
| 134 | MMP1 | NA | MMP1 Entrez, Source | matrix metallopeptidase 1 (interstitial collagenase) | 27012 | -3.065 | -0.1188 | Yes |
| 135 | PDE2A | NA | PDE2A Entrez, Source | phosphodiesterase 2A, cGMP-stimulated | 27080 | -3.285 | -0.0969 | Yes |
| 136 | NAV3 | NA | NAV3 Entrez, Source | neuron navigator 3 | 27085 | -3.304 | -0.0726 | Yes |
| 137 | ISG20 | NA | ISG20 Entrez, Source | interferon stimulated exonuclease gene 20kDa | 27089 | -3.317 | -0.0481 | Yes |
| 138 | CXCL5 | NA | CXCL5 Entrez, Source | chemokine (C-X-C motif) ligand 5 | 27183 | -3.696 | -0.0242 | Yes |
| 139 | FAM155B | NA | 27228 | -3.985 | 0.0037 | Yes |