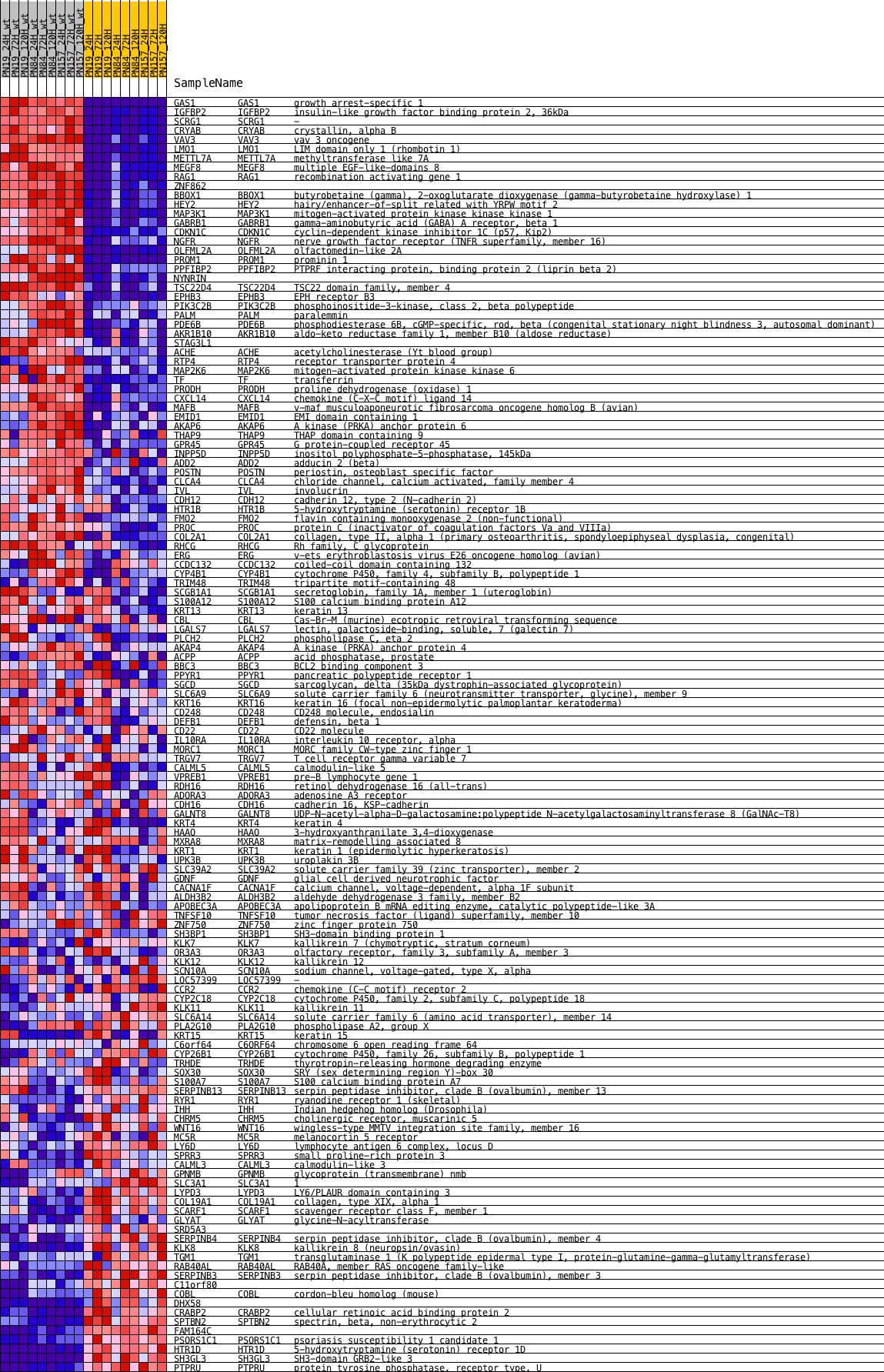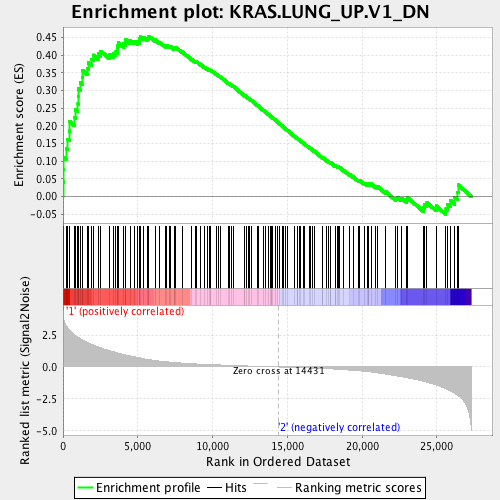
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | KRAS.LUNG_UP.V1_DN |
| Enrichment Score (ES) | 0.4538903 |
| Normalized Enrichment Score (NES) | 1.7478865 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0047248383 |
| FWER p-Value | 0.014 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GAS1 | NA | GAS1 Entrez, Source | growth arrest-specific 1 | 1 | 4.318 | 0.0414 | Yes |
| 2 | IGFBP2 | NA | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 37 | 3.744 | 0.0760 | Yes |
| 3 | SCRG1 | NA | SCRG1 Entrez, Source | - | 50 | 3.656 | 0.1107 | Yes |
| 4 | CRYAB | NA | CRYAB Entrez, Source | crystallin, alpha B | 203 | 3.264 | 0.1364 | Yes |
| 5 | VAV3 | NA | VAV3 Entrez, Source | vav 3 oncogene | 319 | 3.047 | 0.1614 | Yes |
| 6 | LMO1 | NA | LMO1 Entrez, Source | LIM domain only 1 (rhombotin 1) | 414 | 2.907 | 0.1858 | Yes |
| 7 | METTL7A | NA | METTL7A Entrez, Source | methyltransferase like 7A | 433 | 2.889 | 0.2129 | Yes |
| 8 | MEGF8 | NA | MEGF8 Entrez, Source | multiple EGF-like-domains 8 | 786 | 2.489 | 0.2238 | Yes |
| 9 | RAG1 | NA | RAG1 Entrez, Source | recombination activating gene 1 | 842 | 2.440 | 0.2452 | Yes |
| 10 | ZNF862 | NA | 992 | 2.320 | 0.2620 | Yes | ||
| 11 | BBOX1 | NA | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 1012 | 2.310 | 0.2835 | Yes |
| 12 | HEY2 | NA | HEY2 Entrez, Source | hairy/enhancer-of-split related with YRPW motif 2 | 1019 | 2.306 | 0.3054 | Yes |
| 13 | MAP3K1 | NA | MAP3K1 Entrez, Source | mitogen-activated protein kinase kinase kinase 1 | 1139 | 2.220 | 0.3223 | Yes |
| 14 | GABRB1 | NA | GABRB1 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, beta 1 | 1274 | 2.119 | 0.3377 | Yes |
| 15 | CDKN1C | NA | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 1317 | 2.089 | 0.3562 | Yes |
| 16 | NGFR | NA | NGFR Entrez, Source | nerve growth factor receptor (TNFR superfamily, member 16) | 1617 | 1.912 | 0.3636 | Yes |
| 17 | OLFML2A | NA | OLFML2A Entrez, Source | olfactomedin-like 2A | 1700 | 1.871 | 0.3785 | Yes |
| 18 | PROM1 | NA | PROM1 Entrez, Source | prominin 1 | 1895 | 1.778 | 0.3884 | Yes |
| 19 | PPFIBP2 | NA | PPFIBP2 Entrez, Source | PTPRF interacting protein, binding protein 2 (liprin beta 2) | 2015 | 1.716 | 0.4005 | Yes |
| 20 | NYNRIN | NA | 2340 | 1.580 | 0.4038 | Yes | ||
| 21 | TSC22D4 | NA | TSC22D4 Entrez, Source | TSC22 domain family, member 4 | 2516 | 1.500 | 0.4117 | Yes |
| 22 | EPHB3 | NA | EPHB3 Entrez, Source | EPH receptor B3 | 3129 | 1.265 | 0.4013 | Yes |
| 23 | PIK3C2B | NA | PIK3C2B Entrez, Source | phosphoinositide-3-kinase, class 2, beta polypeptide | 3375 | 1.192 | 0.4038 | Yes |
| 24 | PALM | NA | PALM Entrez, Source | paralemmin | 3528 | 1.145 | 0.4092 | Yes |
| 25 | PDE6B | NA | PDE6B Entrez, Source | phosphodiesterase 6B, cGMP-specific, rod, beta (congenital stationary night blindness 3, autosomal dominant) | 3624 | 1.108 | 0.4163 | Yes |
| 26 | AKR1B10 | NA | AKR1B10 Entrez, Source | aldo-keto reductase family 1, member B10 (aldose reductase) | 3645 | 1.100 | 0.4261 | Yes |
| 27 | STAG3L1 | NA | 3688 | 1.085 | 0.4350 | Yes | ||
| 28 | ACHE | NA | ACHE Entrez, Source | acetylcholinesterase (Yt blood group) | 4029 | 0.978 | 0.4319 | Yes |
| 29 | RTP4 | NA | RTP4 Entrez, Source | receptor transporter protein 4 | 4159 | 0.942 | 0.4362 | Yes |
| 30 | MAP2K6 | NA | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 4200 | 0.932 | 0.4436 | Yes |
| 31 | TF | NA | TF Entrez, Source | transferrin | 4501 | 0.861 | 0.4409 | Yes |
| 32 | PRODH | NA | PRODH Entrez, Source | proline dehydrogenase (oxidase) 1 | 4744 | 0.802 | 0.4397 | Yes |
| 33 | CXCL14 | NA | CXCL14 Entrez, Source | chemokine (C-X-C motif) ligand 14 | 4987 | 0.746 | 0.4379 | Yes |
| 34 | MAFB | NA | MAFB Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) | 5115 | 0.716 | 0.4401 | Yes |
| 35 | EMID1 | NA | EMID1 Entrez, Source | EMI domain containing 1 | 5134 | 0.712 | 0.4463 | Yes |
| 36 | AKAP6 | NA | AKAP6 Entrez, Source | A kinase (PRKA) anchor protein 6 | 5156 | 0.707 | 0.4523 | Yes |
| 37 | THAP9 | NA | THAP9 Entrez, Source | THAP domain containing 9 | 5355 | 0.649 | 0.4512 | Yes |
| 38 | GPR45 | NA | GPR45 Entrez, Source | G protein-coupled receptor 45 | 5620 | 0.594 | 0.4472 | Yes |
| 39 | INPP5D | NA | INPP5D Entrez, Source | inositol polyphosphate-5-phosphatase, 145kDa | 5714 | 0.576 | 0.4493 | Yes |
| 40 | ADD2 | NA | ADD2 Entrez, Source | adducin 2 (beta) | 5740 | 0.572 | 0.4539 | Yes |
| 41 | POSTN | NA | POSTN Entrez, Source | periostin, osteoblast specific factor | 6160 | 0.503 | 0.4433 | No |
| 42 | CLCA4 | NA | CLCA4 Entrez, Source | chloride channel, calcium activated, family member 4 | 6461 | 0.457 | 0.4367 | No |
| 43 | IVL | NA | IVL Entrez, Source | involucrin | 6836 | 0.415 | 0.4269 | No |
| 44 | CDH12 | NA | CDH12 Entrez, Source | cadherin 12, type 2 (N-cadherin 2) | 6932 | 0.404 | 0.4273 | No |
| 45 | HTR1B | NA | HTR1B Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 1B | 7098 | 0.386 | 0.4249 | No |
| 46 | FMO2 | NA | FMO2 Entrez, Source | flavin containing monooxygenase 2 (non-functional) | 7213 | 0.374 | 0.4243 | No |
| 47 | PROC | NA | PROC Entrez, Source | protein C (inactivator of coagulation factors Va and VIIIa) | 7423 | 0.352 | 0.4200 | No |
| 48 | COL2A1 | NA | COL2A1 Entrez, Source | collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 7453 | 0.349 | 0.4223 | No |
| 49 | RHCG | NA | RHCG Entrez, Source | Rh family, C glycoprotein | 7531 | 0.341 | 0.4227 | No |
| 50 | ERG | NA | ERG Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog (avian) | 7955 | 0.304 | 0.4101 | No |
| 51 | CCDC132 | NA | CCDC132 Entrez, Source | coiled-coil domain containing 132 | 8607 | 0.256 | 0.3886 | No |
| 52 | CYP4B1 | NA | CYP4B1 Entrez, Source | cytochrome P450, family 4, subfamily B, polypeptide 1 | 8869 | 0.238 | 0.3813 | No |
| 53 | TRIM48 | NA | TRIM48 Entrez, Source | tripartite motif-containing 48 | 8914 | 0.235 | 0.3819 | No |
| 54 | SCGB1A1 | NA | SCGB1A1 Entrez, Source | secretoglobin, family 1A, member 1 (uteroglobin) | 9216 | 0.218 | 0.3729 | No |
| 55 | S100A12 | NA | S100A12 Entrez, Source | S100 calcium binding protein A12 | 9458 | 0.206 | 0.3661 | No |
| 56 | KRT13 | NA | KRT13 Entrez, Source | keratin 13 | 9639 | 0.195 | 0.3613 | No |
| 57 | CBL | NA | CBL Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence | 9769 | 0.190 | 0.3584 | No |
| 58 | LGALS7 | NA | LGALS7 Entrez, Source | lectin, galactoside-binding, soluble, 7 (galectin 7) | 9838 | 0.186 | 0.3577 | No |
| 59 | PLCH2 | NA | PLCH2 Entrez, Source | phospholipase C, eta 2 | 9884 | 0.184 | 0.3578 | No |
| 60 | AKAP4 | NA | AKAP4 Entrez, Source | A kinase (PRKA) anchor protein 4 | 10239 | 0.168 | 0.3464 | No |
| 61 | ACPP | NA | ACPP Entrez, Source | acid phosphatase, prostate | 10414 | 0.160 | 0.3415 | No |
| 62 | BBC3 | NA | BBC3 Entrez, Source | BCL2 binding component 3 | 10553 | 0.153 | 0.3379 | No |
| 63 | PPYR1 | NA | PPYR1 Entrez, Source | pancreatic polypeptide receptor 1 | 11079 | 0.128 | 0.3198 | No |
| 64 | SGCD | NA | SGCD Entrez, Source | sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) | 11107 | 0.127 | 0.3200 | No |
| 65 | SLC6A9 | NA | SLC6A9 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 11257 | 0.120 | 0.3157 | No |
| 66 | KRT16 | NA | KRT16 Entrez, Source | keratin 16 (focal non-epidermolytic palmoplantar keratoderma) | 11408 | 0.113 | 0.3113 | No |
| 67 | CD248 | NA | CD248 Entrez, Source | CD248 molecule, endosialin | 12102 | 0.087 | 0.2866 | No |
| 68 | DEFB1 | NA | DEFB1 Entrez, Source | defensin, beta 1 | 12165 | 0.084 | 0.2852 | No |
| 69 | CD22 | NA | CD22 Entrez, Source | CD22 molecule | 12251 | 0.081 | 0.2828 | No |
| 70 | IL10RA | NA | IL10RA Entrez, Source | interleukin 10 receptor, alpha | 12399 | 0.075 | 0.2781 | No |
| 71 | MORC1 | NA | MORC1 Entrez, Source | MORC family CW-type zinc finger 1 | 12481 | 0.072 | 0.2758 | No |
| 72 | TRGV7 | NA | TRGV7 Entrez, Source | T cell receptor gamma variable 7 | 12575 | 0.068 | 0.2731 | No |
| 73 | CALML5 | NA | CALML5 Entrez, Source | calmodulin-like 5 | 13006 | 0.053 | 0.2578 | No |
| 74 | VPREB1 | NA | VPREB1 Entrez, Source | pre-B lymphocyte gene 1 | 13041 | 0.051 | 0.2570 | No |
| 75 | RDH16 | NA | RDH16 Entrez, Source | retinol dehydrogenase 16 (all-trans) | 13401 | 0.037 | 0.2442 | No |
| 76 | ADORA3 | NA | ADORA3 Entrez, Source | adenosine A3 receptor | 13509 | 0.033 | 0.2405 | No |
| 77 | CDH16 | NA | CDH16 Entrez, Source | cadherin 16, KSP-cadherin | 13744 | 0.024 | 0.2322 | No |
| 78 | GALNT8 | NA | GALNT8 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8) | 13897 | 0.018 | 0.2267 | No |
| 79 | KRT4 | NA | KRT4 Entrez, Source | keratin 4 | 13946 | 0.017 | 0.2251 | No |
| 80 | HAAO | NA | HAAO Entrez, Source | 3-hydroxyanthranilate 3,4-dioxygenase | 13959 | 0.017 | 0.2249 | No |
| 81 | MXRA8 | NA | MXRA8 Entrez, Source | matrix-remodelling associated 8 | 14008 | 0.015 | 0.2232 | No |
| 82 | KRT1 | NA | KRT1 Entrez, Source | keratin 1 (epidermolytic hyperkeratosis) | 14206 | 0.008 | 0.2161 | No |
| 83 | UPK3B | NA | UPK3B Entrez, Source | uroplakin 3B | 14339 | 0.003 | 0.2112 | No |
| 84 | SLC39A2 | NA | SLC39A2 Entrez, Source | solute carrier family 39 (zinc transporter), member 2 | 14483 | -0.002 | 0.2060 | No |
| 85 | GDNF | NA | GDNF Entrez, Source | glial cell derived neurotrophic factor | 14707 | -0.010 | 0.1979 | No |
| 86 | CACNA1F | NA | CACNA1F Entrez, Source | calcium channel, voltage-dependent, alpha 1F subunit | 14776 | -0.012 | 0.1955 | No |
| 87 | ALDH3B2 | NA | ALDH3B2 Entrez, Source | aldehyde dehydrogenase 3 family, member B2 | 14888 | -0.016 | 0.1916 | No |
| 88 | APOBEC3A | NA | APOBEC3A Entrez, Source | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A | 15006 | -0.021 | 0.1875 | No |
| 89 | TNFSF10 | NA | TNFSF10 Entrez, Source | tumor necrosis factor (ligand) superfamily, member 10 | 15485 | -0.040 | 0.1703 | No |
| 90 | ZNF750 | NA | ZNF750 Entrez, Source | zinc finger protein 750 | 15668 | -0.047 | 0.1641 | No |
| 91 | SH3BP1 | NA | SH3BP1 Entrez, Source | SH3-domain binding protein 1 | 15694 | -0.048 | 0.1636 | No |
| 92 | KLK7 | NA | KLK7 Entrez, Source | kallikrein 7 (chymotryptic, stratum corneum) | 15820 | -0.053 | 0.1595 | No |
| 93 | OR3A3 | NA | OR3A3 Entrez, Source | olfactory receptor, family 3, subfamily A, member 3 | 15866 | -0.054 | 0.1584 | No |
| 94 | KLK12 | NA | KLK12 Entrez, Source | kallikrein 12 | 16052 | -0.062 | 0.1522 | No |
| 95 | SCN10A | NA | SCN10A Entrez, Source | sodium channel, voltage-gated, type X, alpha | 16173 | -0.068 | 0.1484 | No |
| 96 | LOC57399 | NA | LOC57399 Entrez, Source | - | 16453 | -0.079 | 0.1389 | No |
| 97 | CCR2 | NA | CCR2 Entrez, Source | chemokine (C-C motif) receptor 2 | 16547 | -0.083 | 0.1363 | No |
| 98 | CYP2C18 | NA | CYP2C18 Entrez, Source | cytochrome P450, family 2, subfamily C, polypeptide 18 | 16706 | -0.089 | 0.1313 | No |
| 99 | KLK11 | NA | KLK11 Entrez, Source | kallikrein 11 | 16791 | -0.093 | 0.1291 | No |
| 100 | SLC6A14 | NA | SLC6A14 Entrez, Source | solute carrier family 6 (amino acid transporter), member 14 | 17324 | -0.116 | 0.1107 | No |
| 101 | PLA2G10 | NA | PLA2G10 Entrez, Source | phospholipase A2, group X | 17383 | -0.119 | 0.1097 | No |
| 102 | KRT15 | NA | KRT15 Entrez, Source | keratin 15 | 17642 | -0.131 | 0.1015 | No |
| 103 | C6orf64 | NA | C6ORF64 Entrez, Source | chromosome 6 open reading frame 64 | 17783 | -0.139 | 0.0976 | No |
| 104 | CYP26B1 | NA | CYP26B1 Entrez, Source | cytochrome P450, family 26, subfamily B, polypeptide 1 | 17875 | -0.145 | 0.0957 | No |
| 105 | TRHDE | NA | TRHDE Entrez, Source | thyrotropin-releasing hormone degrading enzyme | 17887 | -0.146 | 0.0967 | No |
| 106 | SOX30 | NA | SOX30 Entrez, Source | SRY (sex determining region Y)-box 30 | 18192 | -0.165 | 0.0871 | No |
| 107 | S100A7 | NA | S100A7 Entrez, Source | S100 calcium binding protein A7 | 18221 | -0.167 | 0.0877 | No |
| 108 | SERPINB13 | NA | SERPINB13 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 13 | 18338 | -0.174 | 0.0851 | No |
| 109 | RYR1 | NA | RYR1 Entrez, Source | ryanodine receptor 1 (skeletal) | 18403 | -0.178 | 0.0844 | No |
| 110 | IHH | NA | IHH Entrez, Source | Indian hedgehog homolog (Drosophila) | 18476 | -0.184 | 0.0835 | No |
| 111 | CHRM5 | NA | CHRM5 Entrez, Source | cholinergic receptor, muscarinic 5 | 18788 | -0.209 | 0.0741 | No |
| 112 | WNT16 | NA | WNT16 Entrez, Source | wingless-type MMTV integration site family, member 16 | 19151 | -0.239 | 0.0631 | No |
| 113 | MC5R | NA | MC5R Entrez, Source | melanocortin 5 receptor | 19421 | -0.262 | 0.0557 | No |
| 114 | LY6D | NA | LY6D Entrez, Source | lymphocyte antigen 6 complex, locus D | 19787 | -0.298 | 0.0452 | No |
| 115 | SPRR3 | NA | SPRR3 Entrez, Source | small proline-rich protein 3 | 19850 | -0.305 | 0.0458 | No |
| 116 | CALML3 | NA | CALML3 Entrez, Source | calmodulin-like 3 | 20178 | -0.340 | 0.0370 | No |
| 117 | GPNMB | NA | GPNMB Entrez, Source | glycoprotein (transmembrane) nmb | 20342 | -0.361 | 0.0345 | No |
| 118 | SLC3A1 | NA | SLC3A1 Entrez, Source | 1 | 20428 | -0.371 | 0.0349 | No |
| 119 | LYPD3 | NA | LYPD3 Entrez, Source | LY6/PLAUR domain containing 3 | 20456 | -0.375 | 0.0375 | No |
| 120 | COL19A1 | NA | COL19A1 Entrez, Source | collagen, type XIX, alpha 1 | 20601 | -0.393 | 0.0360 | No |
| 121 | SCARF1 | NA | SCARF1 Entrez, Source | scavenger receptor class F, member 1 | 20922 | -0.440 | 0.0285 | No |
| 122 | GLYAT | NA | GLYAT Entrez, Source | glycine-N-acyltransferase | 21048 | -0.460 | 0.0283 | No |
| 123 | SRD5A3 | NA | 21591 | -0.551 | 0.0136 | No | ||
| 124 | SERPINB4 | NA | SERPINB4 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 4 | 22268 | -0.681 | -0.0047 | No |
| 125 | KLK8 | NA | KLK8 Entrez, Source | kallikrein 8 (neuropsin/ovasin) | 22369 | -0.699 | -0.0016 | No |
| 126 | TGM1 | NA | TGM1 Entrez, Source | transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) | 22653 | -0.766 | -0.0047 | No |
| 127 | RAB40AL | NA | RAB40AL Entrez, Source | RAB40A, member RAS oncogene family-like | 22977 | -0.836 | -0.0086 | No |
| 128 | SERPINB3 | NA | SERPINB3 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 3 | 23022 | -0.843 | -0.0021 | No |
| 129 | C11orf80 | NA | 24107 | -1.122 | -0.0312 | No | ||
| 130 | COBL | NA | COBL Entrez, Source | cordon-bleu homolog (mouse) | 24195 | -1.147 | -0.0234 | No |
| 131 | DHX58 | NA | 24343 | -1.193 | -0.0173 | No | ||
| 132 | CRABP2 | NA | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 24956 | -1.400 | -0.0264 | No |
| 133 | SPTBN2 | NA | SPTBN2 Entrez, Source | spectrin, beta, non-erythrocytic 2 | 25615 | -1.695 | -0.0344 | No |
| 134 | FAM164C | NA | 25732 | -1.754 | -0.0218 | No | ||
| 135 | PSORS1C1 | NA | PSORS1C1 Entrez, Source | psoriasis susceptibility 1 candidate 1 | 25902 | -1.858 | -0.0102 | No |
| 136 | HTR1D | NA | HTR1D Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 1D | 26209 | -2.065 | -0.0016 | No |
| 137 | SH3GL3 | NA | SH3GL3 Entrez, Source | SH3-domain GRB2-like 3 | 26398 | -2.214 | 0.0127 | No |
| 138 | PTPRU | NA | PTPRU Entrez, Source | protein tyrosine phosphatase, receptor type, U | 26431 | -2.240 | 0.0330 | No |