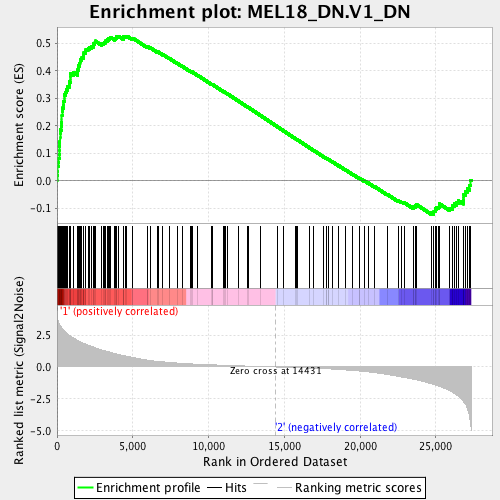
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 1 |
| GeneSet | MEL18_DN.V1_DN |
| Enrichment Score (ES) | 0.52681965 |
| Normalized Enrichment Score (NES) | 1.6762763 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00679783 |
| FWER p-Value | 0.041 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FOS | NA | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 3 | 4.177 | 0.0191 | Yes |
| 2 | COL9A3 | NA | COL9A3 Entrez, Source | collagen, type IX, alpha 3 | 38 | 3.730 | 0.0350 | Yes |
| 3 | MAF | NA | MAF Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | 46 | 3.693 | 0.0517 | Yes |
| 4 | B3GAT1 | NA | B3GAT1 Entrez, Source | beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) | 71 | 3.570 | 0.0672 | Yes |
| 5 | C1orf54 | NA | C1ORF54 Entrez, Source | chromosome 1 open reading frame 54 | 85 | 3.528 | 0.0830 | Yes |
| 6 | PPARGC1A | NA | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 136 | 3.387 | 0.0967 | Yes |
| 7 | PDE4B | NA | PDE4B Entrez, Source | phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | 147 | 3.365 | 0.1118 | Yes |
| 8 | DOK5 | NA | DOK5 Entrez, Source | docking protein 5 | 153 | 3.346 | 0.1270 | Yes |
| 9 | ID4 | NA | ID4 Entrez, Source | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | 172 | 3.304 | 0.1415 | Yes |
| 10 | SYT17 | NA | SYT17 Entrez, Source | synaptotagmin XVII | 190 | 3.284 | 0.1560 | Yes |
| 11 | PLCB1 | NA | PLCB1 Entrez, Source | phospholipase C, beta 1 (phosphoinositide-specific) | 193 | 3.282 | 0.1710 | Yes |
| 12 | PNMAL1 | NA | 200 | 3.272 | 0.1858 | Yes | ||
| 13 | BAALC | NA | BAALC Entrez, Source | brain and acute leukemia, cytoplasmic | 267 | 3.141 | 0.1978 | Yes |
| 14 | HOXB13 | NA | HOXB13 Entrez, Source | homeobox B13 | 288 | 3.103 | 0.2113 | Yes |
| 15 | VAV3 | NA | VAV3 Entrez, Source | vav 3 oncogene | 319 | 3.047 | 0.2242 | Yes |
| 16 | NDRG2 | NA | NDRG2 Entrez, Source | NDRG family member 2 | 320 | 3.045 | 0.2382 | Yes |
| 17 | GULP1 | NA | GULP1 Entrez, Source | GULP, engulfment adaptor PTB domain containing 1 | 330 | 3.029 | 0.2518 | Yes |
| 18 | SOX2 | NA | SOX2 Entrez, Source | SRY (sex determining region Y)-box 2 | 342 | 3.009 | 0.2652 | Yes |
| 19 | PDE7B | NA | PDE7B Entrez, Source | phosphodiesterase 7B | 411 | 2.910 | 0.2761 | Yes |
| 20 | CLIP3 | NA | 421 | 2.902 | 0.2891 | Yes | ||
| 21 | SEPP1 | NA | SEPP1 Entrez, Source | selenoprotein P, plasma, 1 | 479 | 2.830 | 0.3000 | Yes |
| 22 | ID2 | NA | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 481 | 2.829 | 0.3130 | Yes |
| 23 | RGS2 | NA | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 561 | 2.704 | 0.3225 | Yes |
| 24 | EYA4 | NA | EYA4 Entrez, Source | eyes absent homolog 4 (Drosophila) | 612 | 2.664 | 0.3329 | Yes |
| 25 | GTF2I | NA | GTF2I Entrez, Source | general transcription factor II, i | 689 | 2.587 | 0.3420 | Yes |
| 26 | MPPED2 | NA | MPPED2 Entrez, Source | metallophosphoesterase domain containing 2 | 794 | 2.479 | 0.3496 | Yes |
| 27 | GAS2 | NA | GAS2 Entrez, Source | growth arrest-specific 2 | 810 | 2.465 | 0.3603 | Yes |
| 28 | UNC5C | NA | UNC5C Entrez, Source | unc-5 homolog C (C. elegans) | 878 | 2.408 | 0.3689 | Yes |
| 29 | EPHA3 | NA | EPHA3 Entrez, Source | EPH receptor A3 | 880 | 2.407 | 0.3800 | Yes |
| 30 | PEG3 | NA | PEG3 Entrez, Source | paternally expressed 3 | 886 | 2.400 | 0.3908 | Yes |
| 31 | PDLIM5 | NA | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 1063 | 2.270 | 0.3948 | Yes |
| 32 | TMOD1 | NA | TMOD1 Entrez, Source | tropomodulin 1 | 1335 | 2.081 | 0.3944 | Yes |
| 33 | ZCCHC24 | NA | 1343 | 2.076 | 0.4037 | Yes | ||
| 34 | SLC16A4 | NA | SLC16A4 Entrez, Source | solute carrier family 16, member 4 (monocarboxylic acid transporter 5) | 1412 | 2.026 | 0.4105 | Yes |
| 35 | BST2 | NA | BST2 Entrez, Source | bone marrow stromal cell antigen 2 | 1430 | 2.017 | 0.4191 | Yes |
| 36 | KCNJ16 | NA | KCNJ16 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 16 | 1471 | 1.995 | 0.4268 | Yes |
| 37 | ATXN3 | NA | ATXN3 Entrez, Source | ataxin 3 | 1510 | 1.972 | 0.4345 | Yes |
| 38 | SH3BGRL | NA | SH3BGRL Entrez, Source | SH3 domain binding glutamic acid-rich protein like | 1527 | 1.962 | 0.4429 | Yes |
| 39 | WFDC2 | NA | WFDC2 Entrez, Source | WAP four-disulfide core domain 2 | 1637 | 1.902 | 0.4476 | Yes |
| 40 | CA2 | NA | CA2 Entrez, Source | carbonic anhydrase II | 1735 | 1.854 | 0.4526 | Yes |
| 41 | PLEKHA4 | NA | PLEKHA4 Entrez, Source | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 | 1763 | 1.843 | 0.4601 | Yes |
| 42 | CHN2 | NA | CHN2 Entrez, Source | chimerin (chimaerin) 2 | 1770 | 1.842 | 0.4683 | Yes |
| 43 | PROM1 | NA | PROM1 Entrez, Source | prominin 1 | 1895 | 1.778 | 0.4719 | Yes |
| 44 | APOE | NA | APOE Entrez, Source | apolipoprotein E | 1901 | 1.776 | 0.4799 | Yes |
| 45 | GNG7 | NA | GNG7 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 7 | 2040 | 1.702 | 0.4826 | Yes |
| 46 | HIST1H4H | NA | HIST1H4H Entrez, Source | histone cluster 1, H4h | 2133 | 1.663 | 0.4869 | Yes |
| 47 | AK1 | NA | AK1 Entrez, Source | adenylate kinase 1 | 2256 | 1.615 | 0.4898 | Yes |
| 48 | EFHD1 | NA | EFHD1 Entrez, Source | EF-hand domain family, member D1 | 2376 | 1.561 | 0.4926 | Yes |
| 49 | FAM46A | NA | FAM46A Entrez, Source | family with sequence similarity 46, member A | 2390 | 1.553 | 0.4993 | Yes |
| 50 | TMEM176B | NA | TMEM176B Entrez, Source | transmembrane protein 176B | 2498 | 1.506 | 0.5023 | Yes |
| 51 | CRABP1 | NA | CRABP1 Entrez, Source | cellular retinoic acid binding protein 1 | 2500 | 1.506 | 0.5092 | Yes |
| 52 | PID1 | NA | 2913 | 1.345 | 0.5002 | Yes | ||
| 53 | CREM | NA | CREM Entrez, Source | cAMP responsive element modulator | 3043 | 1.296 | 0.5014 | Yes |
| 54 | PCDH8 | NA | PCDH8 Entrez, Source | protocadherin 8 | 3119 | 1.269 | 0.5045 | Yes |
| 55 | SNCAIP | NA | SNCAIP Entrez, Source | synuclein, alpha interacting protein (synphilin) | 3198 | 1.241 | 0.5073 | Yes |
| 56 | ALDH6A1 | NA | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 3221 | 1.234 | 0.5122 | Yes |
| 57 | DIRAS3 | NA | DIRAS3 Entrez, Source | DIRAS family, GTP-binding RAS-like 3 | 3323 | 1.208 | 0.5140 | Yes |
| 58 | PIK3C2B | NA | PIK3C2B Entrez, Source | phosphoinositide-3-kinase, class 2, beta polypeptide | 3375 | 1.192 | 0.5176 | Yes |
| 59 | FAM115A | NA | 3470 | 1.163 | 0.5195 | Yes | ||
| 60 | GAS7 | NA | GAS7 Entrez, Source | growth arrest-specific 7 | 3520 | 1.148 | 0.5229 | Yes |
| 61 | SKAP2 | NA | SKAP2 Entrez, Source | src kinase associated phosphoprotein 2 | 3810 | 1.047 | 0.5171 | Yes |
| 62 | TRIM2 | NA | TRIM2 Entrez, Source | tripartite motif-containing 2 | 3879 | 1.024 | 0.5193 | Yes |
| 63 | TRAPPC6A | NA | TRAPPC6A Entrez, Source | trafficking protein particle complex 6A | 3925 | 1.011 | 0.5223 | Yes |
| 64 | NMNAT2 | NA | NMNAT2 Entrez, Source | nicotinamide nucleotide adenylyltransferase 2 | 3931 | 1.008 | 0.5268 | Yes |
| 65 | PCGF2 | NA | PCGF2 Entrez, Source | polycomb group ring finger 2 | 4071 | 0.967 | 0.5261 | Yes |
| 66 | IFITM1 | NA | IFITM1 Entrez, Source | interferon induced transmembrane protein 1 (9-27) | 4366 | 0.891 | 0.5194 | Yes |
| 67 | CRIP1 | NA | CRIP1 Entrez, Source | cysteine-rich protein 1 (intestinal) | 4388 | 0.885 | 0.5227 | Yes |
| 68 | TMEM176A | NA | TMEM176A Entrez, Source | transmembrane protein 176A | 4393 | 0.884 | 0.5266 | Yes |
| 69 | SEMA3E | NA | SEMA3E Entrez, Source | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E | 4517 | 0.857 | 0.5260 | Yes |
| 70 | PCP4 | NA | PCP4 Entrez, Source | Purkinje cell protein 4 | 4601 | 0.842 | 0.5268 | Yes |
| 71 | SERPINI1 | NA | SERPINI1 Entrez, Source | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 4947 | 0.755 | 0.5176 | No |
| 72 | CXCL14 | NA | CXCL14 Entrez, Source | chemokine (C-X-C motif) ligand 14 | 4987 | 0.746 | 0.5196 | No |
| 73 | KIAA1310 | NA | 5950 | 0.537 | 0.4867 | No | ||
| 74 | GPD1L | NA | GPD1L Entrez, Source | glycerol-3-phosphate dehydrogenase 1-like | 5970 | 0.533 | 0.4884 | No |
| 75 | A2M | NA | A2M Entrez, Source | alpha-2-macroglobulin | 6145 | 0.506 | 0.4843 | No |
| 76 | DPYD | NA | DPYD Entrez, Source | dihydropyrimidine dehydrogenase | 6599 | 0.441 | 0.4697 | No |
| 77 | VPS8 | NA | VPS8 Entrez, Source | vacuolar protein sorting 8 homolog (S. cerevisiae) | 6663 | 0.433 | 0.4694 | No |
| 78 | MGC2889 | NA | MGC2889 Entrez, Source | - | 6978 | 0.400 | 0.4597 | No |
| 79 | GPR88 | NA | GPR88 Entrez, Source | G protein-coupled receptor 88 | 7382 | 0.357 | 0.4465 | No |
| 80 | GLS2 | NA | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 7913 | 0.308 | 0.4284 | No |
| 81 | RBP4 | NA | RBP4 Entrez, Source | retinol binding protein 4, plasma | 8284 | 0.278 | 0.4161 | No |
| 82 | TCEAL2 | NA | TCEAL2 Entrez, Source | transcription elongation factor A (SII)-like 2 | 8803 | 0.241 | 0.3981 | No |
| 83 | EBF2 | NA | EBF2 Entrez, Source | early B-cell factor 2 | 8871 | 0.238 | 0.3967 | No |
| 84 | ESRP1 | NA | 8900 | 0.236 | 0.3968 | No | ||
| 85 | MAOB | NA | MAOB Entrez, Source | monoamine oxidase B | 9285 | 0.215 | 0.3837 | No |
| 86 | CDH10 | NA | CDH10 Entrez, Source | cadherin 10, type 2 (T2-cadherin) | 10163 | 0.172 | 0.3522 | No |
| 87 | SLPI | NA | SLPI Entrez, Source | secretory leukocyte peptidase inhibitor | 10226 | 0.168 | 0.3507 | No |
| 88 | PLN | NA | PLN Entrez, Source | phospholamban | 10248 | 0.167 | 0.3507 | No |
| 89 | EPB41L3 | NA | EPB41L3 Entrez, Source | erythrocyte membrane protein band 4.1-like 3 | 10988 | 0.133 | 0.3241 | No |
| 90 | DLK1 | NA | DLK1 Entrez, Source | delta-like 1 homolog (Drosophila) | 11014 | 0.132 | 0.3238 | No |
| 91 | BLNK | NA | BLNK Entrez, Source | B-cell linker | 11083 | 0.128 | 0.3219 | No |
| 92 | IFIT2 | NA | IFIT2 Entrez, Source | interferon-induced protein with tetratricopeptide repeats 2 | 11241 | 0.121 | 0.3166 | No |
| 93 | STK17B | NA | STK17B Entrez, Source | serine/threonine kinase 17b (apoptosis-inducing) | 11936 | 0.093 | 0.2915 | No |
| 94 | AQP3 | NA | AQP3 Entrez, Source | aquaporin 3 (Gill blood group) | 12539 | 0.070 | 0.2697 | No |
| 95 | SMR3B | NA | SMR3B Entrez, Source | submaxillary gland androgen regulated protein 3 homolog B (mouse) | 12625 | 0.066 | 0.2669 | No |
| 96 | SCNN1A | NA | SCNN1A Entrez, Source | sodium channel, nonvoltage-gated 1 alpha | 13451 | 0.035 | 0.2367 | No |
| 97 | CUX2 | NA | 14555 | -0.004 | 0.1961 | No | ||
| 98 | COL14A1 | NA | COL14A1 Entrez, Source | collagen, type XIV, alpha 1 (undulin) | 14969 | -0.020 | 0.1810 | No |
| 99 | GTF2A1L | NA | 15724 | -0.049 | 0.1535 | No | ||
| 100 | LECT1 | NA | LECT1 Entrez, Source | leukocyte cell derived chemotaxin 1 | 15777 | -0.051 | 0.1518 | No |
| 101 | SLC17A6 | NA | SLC17A6 Entrez, Source | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 | 15850 | -0.054 | 0.1494 | No |
| 102 | CCKAR | NA | CCKAR Entrez, Source | cholecystokinin A receptor | 16682 | -0.088 | 0.1193 | No |
| 103 | STON1 | NA | STON1 Entrez, Source | stonin 1 | 16900 | -0.097 | 0.1117 | No |
| 104 | IFI27 | NA | IFI27 Entrez, Source | interferon, alpha-inducible protein 27 | 17553 | -0.127 | 0.0883 | No |
| 105 | OLFM4 | NA | OLFM4 Entrez, Source | olfactomedin 4 | 17764 | -0.138 | 0.0812 | No |
| 106 | ACCN1 | NA | ACCN1 Entrez, Source | amiloride-sensitive cation channel 1, neuronal (degenerin) | 17801 | -0.140 | 0.0806 | No |
| 107 | DIO2 | NA | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 17895 | -0.147 | 0.0778 | No |
| 108 | PENK | NA | PENK Entrez, Source | proenkephalin | 17908 | -0.148 | 0.0781 | No |
| 109 | SULF1 | NA | SULF1 Entrez, Source | sulfatase 1 | 18163 | -0.163 | 0.0695 | No |
| 110 | CDH22 | NA | CDH22 Entrez, Source | cadherin-like 22 | 18597 | -0.193 | 0.0544 | No |
| 111 | GPR137B | NA | GPR137B Entrez, Source | G protein-coupled receptor 137B | 19045 | -0.231 | 0.0390 | No |
| 112 | IRF9 | NA | 19523 | -0.272 | 0.0227 | No | ||
| 113 | TSFM | NA | TSFM Entrez, Source | Ts translation elongation factor, mitochondrial | 19929 | -0.313 | 0.0093 | No |
| 114 | SIM1 | NA | SIM1 Entrez, Source | single-minded homolog 1 (Drosophila) | 19952 | -0.315 | 0.0099 | No |
| 115 | MIA | NA | MIA Entrez, Source | melanoma inhibitory activity | 20282 | -0.352 | -0.0006 | No |
| 116 | EGFL6 | NA | EGFL6 Entrez, Source | EGF-like-domain, multiple 6 | 20516 | -0.382 | -0.0074 | No |
| 117 | C16orf80 | NA | C16ORF80 Entrez, Source | chromosome 16 open reading frame 80 | 20949 | -0.445 | -0.0212 | No |
| 118 | PRPF6 | NA | PRPF6 Entrez, Source | PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) | 21779 | -0.587 | -0.0490 | No |
| 119 | DDC | NA | DDC Entrez, Source | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 22510 | -0.732 | -0.0725 | No |
| 120 | FANCF | NA | FANCF Entrez, Source | Fanconi anemia, complementation group F | 22736 | -0.781 | -0.0772 | No |
| 121 | BAG3 | NA | BAG3 Entrez, Source | BCL2-associated athanogene 3 | 22899 | -0.819 | -0.0794 | No |
| 122 | TSPAN1 | NA | TSPAN1 Entrez, Source | tetraspanin 1 | 23502 | -0.951 | -0.0972 | No |
| 123 | NTS | NA | NTS Entrez, Source | neurotensin | 23518 | -0.956 | -0.0933 | No |
| 124 | PTER | NA | PTER Entrez, Source | phosphotriesterase related | 23633 | -0.988 | -0.0930 | No |
| 125 | TFAP2A | NA | TFAP2A Entrez, Source | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | 23640 | -0.991 | -0.0887 | No |
| 126 | PHYH | NA | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 23709 | -1.009 | -0.0865 | No |
| 127 | DMD | NA | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 24692 | -1.306 | -0.1166 | No |
| 128 | UBE2L6 | NA | UBE2L6 Entrez, Source | ubiquitin-conjugating enzyme E2L 6 | 24851 | -1.360 | -0.1162 | No |
| 129 | DDX60 | NA | 24864 | -1.366 | -0.1104 | No | ||
| 130 | DUSP2 | NA | DUSP2 Entrez, Source | dual specificity phosphatase 2 | 24949 | -1.396 | -0.1070 | No |
| 131 | CRABP2 | NA | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 24956 | -1.400 | -0.1008 | No |
| 132 | C9orf46 | NA | C9ORF46 Entrez, Source | chromosome 9 open reading frame 46 | 25030 | -1.431 | -0.0969 | No |
| 133 | OAS1 | NA | OAS1 Entrez, Source | 2',5'-oligoadenylate synthetase 1, 40/46kDa | 25176 | -1.492 | -0.0954 | No |
| 134 | IFIT1 | NA | IFIT1 Entrez, Source | interferon-induced protein with tetratricopeptide repeats 1 | 25217 | -1.508 | -0.0900 | No |
| 135 | MCTS1 | NA | MCTS1 Entrez, Source | malignant T cell amplified sequence 1 | 25232 | -1.514 | -0.0835 | No |
| 136 | IFI6 | NA | IFI6 Entrez, Source | interferon, alpha-inducible protein 6 | 25912 | -1.862 | -0.0999 | No |
| 137 | FRMD4B | NA | FRMD4B Entrez, Source | FERM domain containing 4B | 26086 | -1.973 | -0.0972 | No |
| 138 | MVP | NA | MVP Entrez, Source | major vault protein | 26105 | -1.983 | -0.0888 | No |
| 139 | JUP | NA | JUP Entrez, Source | junction plakoglobin | 26226 | -2.076 | -0.0836 | No |
| 140 | PDE5A | NA | PDE5A Entrez, Source | phosphodiesterase 5A, cGMP-specific | 26369 | -2.191 | -0.0788 | No |
| 141 | EEF1A2 | NA | EEF1A2 Entrez, Source | eukaryotic translation elongation factor 1 alpha 2 | 26472 | -2.272 | -0.0721 | No |
| 142 | OSBPL10 | NA | OSBPL10 Entrez, Source | oxysterol binding protein-like 10 | 26811 | -2.666 | -0.0723 | No |
| 143 | SOCS2 | NA | SOCS2 Entrez, Source | suppressor of cytokine signaling 2 | 26833 | -2.703 | -0.0606 | No |
| 144 | MAP7 | NA | MAP7 Entrez, Source | microtubule-associated protein 7 | 26837 | -2.718 | -0.0483 | No |
| 145 | IL1R1 | NA | IL1R1 Entrez, Source | interleukin 1 receptor, type I | 26964 | -2.933 | -0.0394 | No |
| 146 | PKP2 | NA | PKP2 Entrez, Source | plakophilin 2 | 27054 | -3.194 | -0.0280 | No |
| 147 | LY6E | NA | LY6E Entrez, Source | lymphocyte antigen 6 complex, locus E | 27224 | -3.952 | -0.0161 | No |
| 148 | EGR1 | NA | EGR1 Entrez, Source | early growth response 1 | 27274 | -4.330 | 0.0020 | No |