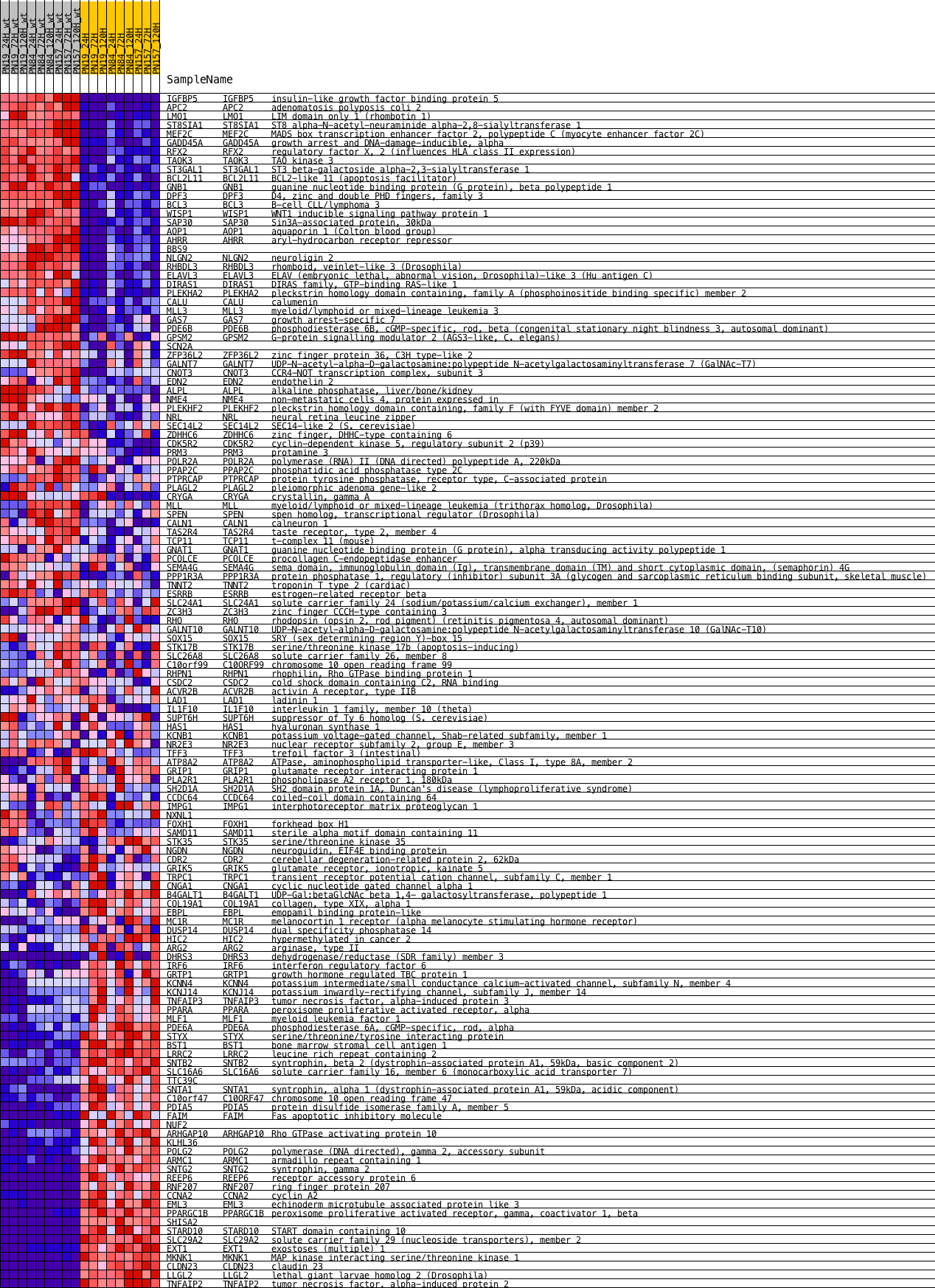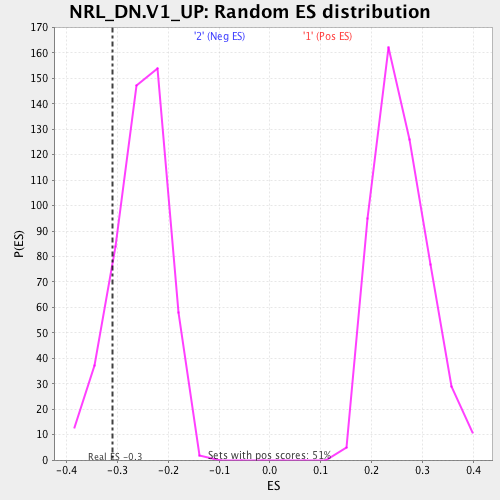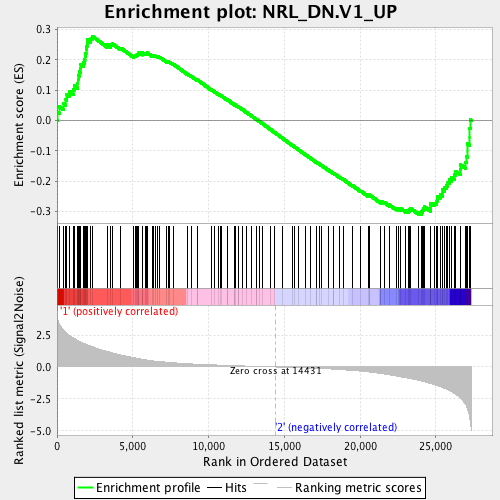
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | NRL_DN.V1_UP |
| Enrichment Score (ES) | -0.30940127 |
| Normalized Enrichment Score (NES) | -1.2117006 |
| Nominal p-value | 0.15353535 |
| FDR q-value | 0.24705233 |
| FWER p-Value | 0.907 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IGFBP5 | NA | IGFBP5 Entrez, Source | insulin-like growth factor binding protein 5 | 10 | 3.987 | 0.0274 | No |
| 2 | APC2 | NA | APC2 Entrez, Source | adenomatosis polyposis coli 2 | 145 | 3.366 | 0.0458 | No |
| 3 | LMO1 | NA | LMO1 Entrez, Source | LIM domain only 1 (rhombotin 1) | 414 | 2.907 | 0.0562 | No |
| 4 | ST8SIA1 | NA | ST8SIA1 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | 577 | 2.693 | 0.0690 | No |
| 5 | MEF2C | NA | MEF2C Entrez, Source | MADS box transcription enhancer factor 2, polypeptide C (myocyte enhancer factor 2C) | 648 | 2.622 | 0.0846 | No |
| 6 | GADD45A | NA | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 793 | 2.479 | 0.0965 | No |
| 7 | RFX2 | NA | RFX2 Entrez, Source | regulatory factor X, 2 (influences HLA class II expression) | 1061 | 2.271 | 0.1025 | No |
| 8 | TAOK3 | NA | TAOK3 Entrez, Source | TAO kinase 3 | 1118 | 2.235 | 0.1160 | No |
| 9 | ST3GAL1 | NA | ST3GAL1 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 1 | 1368 | 2.060 | 0.1212 | No |
| 10 | BCL2L11 | NA | BCL2L11 Entrez, Source | BCL2-like 11 (apoptosis facilitator) | 1377 | 2.056 | 0.1352 | No |
| 11 | GNB1 | NA | GNB1 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 1 | 1385 | 2.051 | 0.1492 | No |
| 12 | DPF3 | NA | DPF3 Entrez, Source | D4, zinc and double PHD fingers, family 3 | 1453 | 2.005 | 0.1606 | No |
| 13 | BCL3 | NA | BCL3 Entrez, Source | B-cell CLL/lymphoma 3 | 1523 | 1.965 | 0.1718 | No |
| 14 | WISP1 | NA | WISP1 Entrez, Source | WNT1 inducible signaling pathway protein 1 | 1568 | 1.941 | 0.1836 | No |
| 15 | SAP30 | NA | SAP30 Entrez, Source | Sin3A-associated protein, 30kDa | 1726 | 1.859 | 0.1908 | No |
| 16 | AQP1 | NA | AQP1 Entrez, Source | aquaporin 1 (Colton blood group) | 1811 | 1.816 | 0.2003 | No |
| 17 | AHRR | NA | AHRR Entrez, Source | aryl-hydrocarbon receptor repressor | 1840 | 1.806 | 0.2119 | No |
| 18 | BBS9 | NA | 1887 | 1.784 | 0.2226 | No | ||
| 19 | NLGN2 | NA | NLGN2 Entrez, Source | neuroligin 2 | 1909 | 1.773 | 0.2341 | No |
| 20 | RHBDL3 | NA | RHBDL3 Entrez, Source | rhomboid, veinlet-like 3 (Drosophila) | 1942 | 1.756 | 0.2452 | No |
| 21 | ELAVL3 | NA | ELAVL3 Entrez, Source | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) | 1976 | 1.738 | 0.2560 | No |
| 22 | DIRAS1 | NA | DIRAS1 Entrez, Source | DIRAS family, GTP-binding RAS-like 1 | 2004 | 1.722 | 0.2670 | No |
| 23 | PLEKHA2 | NA | PLEKHA2 Entrez, Source | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 | 2233 | 1.622 | 0.2699 | No |
| 24 | CALU | NA | CALU Entrez, Source | calumenin | 2307 | 1.592 | 0.2783 | No |
| 25 | MLL3 | NA | MLL3 Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia 3 | 3311 | 1.210 | 0.2498 | No |
| 26 | GAS7 | NA | GAS7 Entrez, Source | growth arrest-specific 7 | 3520 | 1.148 | 0.2502 | No |
| 27 | PDE6B | NA | PDE6B Entrez, Source | phosphodiesterase 6B, cGMP-specific, rod, beta (congenital stationary night blindness 3, autosomal dominant) | 3624 | 1.108 | 0.2541 | No |
| 28 | GPSM2 | NA | GPSM2 Entrez, Source | G-protein signalling modulator 2 (AGS3-like, C. elegans) | 4209 | 0.929 | 0.2390 | No |
| 29 | SCN2A | NA | 5072 | 0.728 | 0.2124 | No | ||
| 30 | ZFP36L2 | NA | ZFP36L2 Entrez, Source | zinc finger protein 36, C3H type-like 2 | 5144 | 0.709 | 0.2147 | No |
| 31 | GALNT7 | NA | GALNT7 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) | 5236 | 0.682 | 0.2161 | No |
| 32 | CNOT3 | NA | CNOT3 Entrez, Source | CCR4-NOT transcription complex, subunit 3 | 5284 | 0.666 | 0.2190 | No |
| 33 | EDN2 | NA | EDN2 Entrez, Source | endothelin 2 | 5342 | 0.651 | 0.2215 | No |
| 34 | ALPL | NA | ALPL Entrez, Source | alkaline phosphatase, liver/bone/kidney | 5400 | 0.638 | 0.2238 | No |
| 35 | NME4 | NA | NME4 Entrez, Source | non-metastatic cells 4, protein expressed in | 5613 | 0.595 | 0.2201 | No |
| 36 | PLEKHF2 | NA | PLEKHF2 Entrez, Source | pleckstrin homology domain containing, family F (with FYVE domain) member 2 | 5646 | 0.588 | 0.2231 | No |
| 37 | NRL | NA | NRL Entrez, Source | neural retina leucine zipper | 5810 | 0.559 | 0.2209 | No |
| 38 | SEC14L2 | NA | SEC14L2 Entrez, Source | SEC14-like 2 (S. cerevisiae) | 5882 | 0.550 | 0.2222 | No |
| 39 | ZDHHC6 | NA | ZDHHC6 Entrez, Source | zinc finger, DHHC-type containing 6 | 5961 | 0.535 | 0.2230 | No |
| 40 | CDK5R2 | NA | CDK5R2 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 2 (p39) | 6323 | 0.478 | 0.2131 | No |
| 41 | PRM3 | NA | PRM3 Entrez, Source | protamine 3 | 6365 | 0.471 | 0.2148 | No |
| 42 | POLR2A | NA | POLR2A Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | 6507 | 0.452 | 0.2128 | No |
| 43 | PPAP2C | NA | PPAP2C Entrez, Source | phosphatidic acid phosphatase type 2C | 6641 | 0.436 | 0.2109 | No |
| 44 | PTPRCAP | NA | PTPRCAP Entrez, Source | protein tyrosine phosphatase, receptor type, C-associated protein | 6762 | 0.422 | 0.2095 | No |
| 45 | PLAGL2 | NA | PLAGL2 Entrez, Source | pleiomorphic adenoma gene-like 2 | 7242 | 0.372 | 0.1944 | No |
| 46 | CRYGA | NA | CRYGA Entrez, Source | crystallin, gamma A | 7327 | 0.362 | 0.1939 | No |
| 47 | MLL | NA | MLL Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | 7425 | 0.351 | 0.1927 | No |
| 48 | SPEN | NA | SPEN Entrez, Source | spen homolog, transcriptional regulator (Drosophila) | 7683 | 0.327 | 0.1855 | No |
| 49 | CALN1 | NA | CALN1 Entrez, Source | calneuron 1 | 8602 | 0.256 | 0.1536 | No |
| 50 | TAS2R4 | NA | TAS2R4 Entrez, Source | taste receptor, type 2, member 4 | 8862 | 0.238 | 0.1457 | No |
| 51 | TCP11 | NA | TCP11 Entrez, Source | t-complex 11 (mouse) | 9233 | 0.217 | 0.1336 | No |
| 52 | GNAT1 | NA | GNAT1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 | 9243 | 0.217 | 0.1348 | No |
| 53 | PCOLCE | NA | PCOLCE Entrez, Source | procollagen C-endopeptidase enhancer | 10193 | 0.170 | 0.1011 | No |
| 54 | SEMA4G | NA | SEMA4G Entrez, Source | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G | 10386 | 0.161 | 0.0951 | No |
| 55 | PPP1R3A | NA | PPP1R3A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 3A (glycogen and sarcoplasmic reticulum binding subunit, skeletal muscle) | 10657 | 0.149 | 0.0862 | No |
| 56 | TNNT2 | NA | TNNT2 Entrez, Source | troponin T type 2 (cardiac) | 10748 | 0.144 | 0.0839 | No |
| 57 | ESRRB | NA | ESRRB Entrez, Source | estrogen-related receptor beta | 10871 | 0.138 | 0.0804 | No |
| 58 | SLC24A1 | NA | SLC24A1 Entrez, Source | solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | 11215 | 0.122 | 0.0686 | No |
| 59 | ZC3H3 | NA | ZC3H3 Entrez, Source | zinc finger CCCH-type containing 3 | 11255 | 0.120 | 0.0680 | No |
| 60 | RHO | NA | RHO Entrez, Source | rhodopsin (opsin 2, rod pigment) (retinitis pigmentosa 4, autosomal dominant) | 11718 | 0.101 | 0.0518 | No |
| 61 | GALNT10 | NA | GALNT10 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) | 11732 | 0.100 | 0.0520 | No |
| 62 | SOX15 | NA | SOX15 Entrez, Source | SRY (sex determining region Y)-box 15 | 11785 | 0.098 | 0.0508 | No |
| 63 | STK17B | NA | STK17B Entrez, Source | serine/threonine kinase 17b (apoptosis-inducing) | 11936 | 0.093 | 0.0459 | No |
| 64 | SLC26A8 | NA | SLC26A8 Entrez, Source | solute carrier family 26, member 8 | 11961 | 0.092 | 0.0456 | No |
| 65 | C10orf99 | NA | C10ORF99 Entrez, Source | chromosome 10 open reading frame 99 | 12203 | 0.082 | 0.0373 | No |
| 66 | RHPN1 | NA | RHPN1 Entrez, Source | rhophilin, Rho GTPase binding protein 1 | 12472 | 0.072 | 0.0280 | No |
| 67 | CSDC2 | NA | CSDC2 Entrez, Source | cold shock domain containing C2, RNA binding | 12814 | 0.060 | 0.0159 | No |
| 68 | ACVR2B | NA | ACVR2B Entrez, Source | activin A receptor, type IIB | 13139 | 0.047 | 0.0043 | No |
| 69 | LAD1 | NA | LAD1 Entrez, Source | ladinin 1 | 13168 | 0.046 | 0.0036 | No |
| 70 | IL1F10 | NA | IL1F10 Entrez, Source | interleukin 1 family, member 10 (theta) | 13360 | 0.039 | -0.0032 | No |
| 71 | SUPT6H | NA | SUPT6H Entrez, Source | suppressor of Ty 6 homolog (S. cerevisiae) | 13565 | 0.030 | -0.0105 | No |
| 72 | HAS1 | NA | HAS1 Entrez, Source | hyaluronan synthase 1 | 14098 | 0.012 | -0.0299 | No |
| 73 | KCNB1 | NA | KCNB1 Entrez, Source | potassium voltage-gated channel, Shab-related subfamily, member 1 | 14340 | 0.003 | -0.0388 | No |
| 74 | NR2E3 | NA | NR2E3 Entrez, Source | nuclear receptor subfamily 2, group E, member 3 | 14893 | -0.017 | -0.0590 | No |
| 75 | TFF3 | NA | TFF3 Entrez, Source | trefoil factor 3 (intestinal) | 15504 | -0.041 | -0.0811 | No |
| 76 | ATP8A2 | NA | ATP8A2 Entrez, Source | ATPase, aminophospholipid transporter-like, Class I, type 8A, member 2 | 15641 | -0.046 | -0.0858 | No |
| 77 | GRIP1 | NA | GRIP1 Entrez, Source | glutamate receptor interacting protein 1 | 15922 | -0.056 | -0.0957 | No |
| 78 | PLA2R1 | NA | PLA2R1 Entrez, Source | phospholipase A2 receptor 1, 180kDa | 16358 | -0.076 | -0.1112 | No |
| 79 | SH2D1A | NA | SH2D1A Entrez, Source | SH2 domain protein 1A, Duncan's disease (lymphoproliferative syndrome) | 16694 | -0.089 | -0.1229 | No |
| 80 | CCDC64 | NA | CCDC64 Entrez, Source | coiled-coil domain containing 64 | 17110 | -0.106 | -0.1374 | No |
| 81 | IMPG1 | NA | IMPG1 Entrez, Source | interphotoreceptor matrix proteoglycan 1 | 17119 | -0.107 | -0.1369 | No |
| 82 | NXNL1 | NA | 17327 | -0.116 | -0.1438 | No | ||
| 83 | FOXH1 | NA | FOXH1 Entrez, Source | forkhead box H1 | 17430 | -0.121 | -0.1467 | No |
| 84 | SAMD11 | NA | SAMD11 Entrez, Source | sterile alpha motif domain containing 11 | 17888 | -0.147 | -0.1624 | No |
| 85 | STK35 | NA | STK35 Entrez, Source | serine/threonine kinase 35 | 18257 | -0.169 | -0.1748 | No |
| 86 | NGDN | NA | NGDN Entrez, Source | neuroguidin, EIF4E binding protein | 18268 | -0.170 | -0.1740 | No |
| 87 | CDR2 | NA | CDR2 Entrez, Source | cerebellar degeneration-related protein 2, 62kDa | 18628 | -0.196 | -0.1858 | No |
| 88 | GRIK5 | NA | GRIK5 Entrez, Source | glutamate receptor, ionotropic, kainate 5 | 18885 | -0.216 | -0.1937 | No |
| 89 | TRPC1 | NA | TRPC1 Entrez, Source | transient receptor potential cation channel, subfamily C, member 1 | 19500 | -0.270 | -0.2144 | No |
| 90 | CNGA1 | NA | CNGA1 Entrez, Source | cyclic nucleotide gated channel alpha 1 | 20048 | -0.326 | -0.2323 | No |
| 91 | B4GALT1 | NA | B4GALT1 Entrez, Source | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 | 20527 | -0.383 | -0.2472 | No |
| 92 | COL19A1 | NA | COL19A1 Entrez, Source | collagen, type XIX, alpha 1 | 20601 | -0.393 | -0.2472 | No |
| 93 | EBPL | NA | EBPL Entrez, Source | emopamil binding protein-like | 20617 | -0.395 | -0.2450 | No |
| 94 | MC1R | NA | MC1R Entrez, Source | melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) | 21346 | -0.506 | -0.2682 | No |
| 95 | DUSP14 | NA | DUSP14 Entrez, Source | dual specificity phosphatase 14 | 21366 | -0.509 | -0.2654 | No |
| 96 | HIC2 | NA | HIC2 Entrez, Source | hypermethylated in cancer 2 | 21620 | -0.555 | -0.2708 | No |
| 97 | ARG2 | NA | ARG2 Entrez, Source | arginase, type II | 21927 | -0.613 | -0.2778 | No |
| 98 | DHRS3 | NA | DHRS3 Entrez, Source | dehydrogenase/reductase (SDR family) member 3 | 22386 | -0.704 | -0.2898 | No |
| 99 | IRF6 | NA | IRF6 Entrez, Source | interferon regulatory factor 6 | 22539 | -0.740 | -0.2902 | No |
| 100 | GRTP1 | NA | GRTP1 Entrez, Source | growth hormone regulated TBC protein 1 | 22683 | -0.770 | -0.2901 | No |
| 101 | KCNN4 | NA | KCNN4 Entrez, Source | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | 23007 | -0.840 | -0.2961 | No |
| 102 | KCNJ14 | NA | KCNJ14 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 14 | 23202 | -0.880 | -0.2972 | No |
| 103 | TNFAIP3 | NA | TNFAIP3 Entrez, Source | tumor necrosis factor, alpha-induced protein 3 | 23231 | -0.887 | -0.2920 | No |
| 104 | PPARA | NA | PPARA Entrez, Source | peroxisome proliferative activated receptor, alpha | 23325 | -0.907 | -0.2891 | No |
| 105 | MLF1 | NA | MLF1 Entrez, Source | myeloid leukemia factor 1 | 23877 | -1.059 | -0.3020 | Yes |
| 106 | PDE6A | NA | PDE6A Entrez, Source | phosphodiesterase 6A, cGMP-specific, rod, alpha | 24050 | -1.104 | -0.3007 | Yes |
| 107 | STYX | NA | STYX Entrez, Source | serine/threonine/tyrosine interacting protein | 24104 | -1.120 | -0.2948 | Yes |
| 108 | BST1 | NA | BST1 Entrez, Source | bone marrow stromal cell antigen 1 | 24154 | -1.136 | -0.2888 | Yes |
| 109 | LRRC2 | NA | LRRC2 Entrez, Source | leucine rich repeat containing 2 | 24238 | -1.161 | -0.2837 | Yes |
| 110 | SNTB2 | NA | SNTB2 Entrez, Source | syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) | 24643 | -1.291 | -0.2896 | Yes |
| 111 | SLC16A6 | NA | SLC16A6 Entrez, Source | solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 24660 | -1.296 | -0.2812 | Yes |
| 112 | TTC39C | NA | 24663 | -1.297 | -0.2722 | Yes | ||
| 113 | SNTA1 | NA | SNTA1 Entrez, Source | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | 24914 | -1.385 | -0.2718 | Yes |
| 114 | C10orf47 | NA | C10ORF47 Entrez, Source | chromosome 10 open reading frame 47 | 25034 | -1.432 | -0.2662 | Yes |
| 115 | PDIA5 | NA | PDIA5 Entrez, Source | protein disulfide isomerase family A, member 5 | 25079 | -1.452 | -0.2578 | Yes |
| 116 | FAIM | NA | FAIM Entrez, Source | Fas apoptotic inhibitory molecule | 25124 | -1.473 | -0.2491 | Yes |
| 117 | NUF2 | NA | 25290 | -1.544 | -0.2445 | Yes | ||
| 118 | ARHGAP10 | NA | ARHGAP10 Entrez, Source | Rho GTPase activating protein 10 | 25435 | -1.610 | -0.2386 | Yes |
| 119 | KLHL36 | NA | 25442 | -1.612 | -0.2276 | Yes | ||
| 120 | POLG2 | NA | POLG2 Entrez, Source | polymerase (DNA directed), gamma 2, accessory subunit | 25575 | -1.677 | -0.2208 | Yes |
| 121 | ARMC1 | NA | ARMC1 Entrez, Source | armadillo repeat containing 1 | 25670 | -1.721 | -0.2123 | Yes |
| 122 | SNTG2 | NA | SNTG2 Entrez, Source | syntrophin, gamma 2 | 25788 | -1.785 | -0.2042 | Yes |
| 123 | REEP6 | NA | REEP6 Entrez, Source | receptor accessory protein 6 | 25886 | -1.850 | -0.1949 | Yes |
| 124 | RNF207 | NA | RNF207 Entrez, Source | ring finger protein 207 | 26043 | -1.945 | -0.1871 | Yes |
| 125 | CCNA2 | NA | CCNA2 Entrez, Source | cyclin A2 | 26210 | -2.066 | -0.1788 | Yes |
| 126 | EML3 | NA | EML3 Entrez, Source | echinoderm microtubule associated protein like 3 | 26296 | -2.126 | -0.1672 | Yes |
| 127 | PPARGC1B | NA | PPARGC1B Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, beta | 26621 | -2.408 | -0.1623 | Yes |
| 128 | SHISA2 | NA | 26634 | -2.422 | -0.1459 | Yes | ||
| 129 | STARD10 | NA | STARD10 Entrez, Source | START domain containing 10 | 26945 | -2.900 | -0.1372 | Yes |
| 130 | SLC29A2 | NA | SLC29A2 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 2 | 26988 | -3.010 | -0.1178 | Yes |
| 131 | EXT1 | NA | EXT1 Entrez, Source | exostoses (multiple) 1 | 27055 | -3.195 | -0.0980 | Yes |
| 132 | MKNK1 | NA | MKNK1 Entrez, Source | MAP kinase interacting serine/threonine kinase 1 | 27056 | -3.195 | -0.0758 | Yes |
| 133 | CLDN23 | NA | CLDN23 Entrez, Source | claudin 23 | 27232 | -4.022 | -0.0542 | Yes |
| 134 | LLGL2 | NA | LLGL2 Entrez, Source | lethal giant larvae homolog 2 (Drosophila) | 27234 | -4.038 | -0.0262 | Yes |
| 135 | TNFAIP2 | NA | TNFAIP2 Entrez, Source | tumor necrosis factor, alpha-induced protein 2 | 27266 | -4.266 | 0.0023 | Yes |