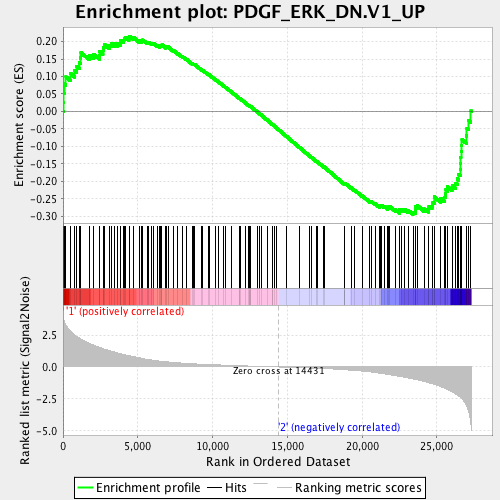
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | PDGF_ERK_DN.V1_UP |
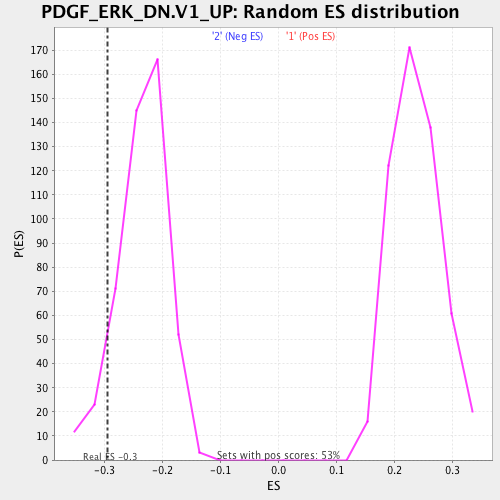
| Enrichment Score (ES) | -0.29501608 |
| Normalized Enrichment Score (NES) | -1.2530729 |
| Nominal p-value | 0.08050848 |
| FDR q-value | 0.2124344 |
| FWER p-Value | 0.845 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PMP22 | NA | PMP22 Entrez, Source | peripheral myelin protein 22 | 41 | 3.713 | 0.0261 | No |
| 2 | MAP2 | NA | MAP2 Entrez, Source | microtubule-associated protein 2 | 54 | 3.643 | 0.0527 | No |
| 3 | TSPAN5 | NA | TSPAN5 Entrez, Source | tetraspanin 5 | 65 | 3.591 | 0.0789 | No |
| 4 | SOX21 | NA | SOX21 Entrez, Source | SRY (sex determining region Y)-box 21 | 189 | 3.287 | 0.0988 | No |
| 5 | CORO2B | NA | CORO2B Entrez, Source | coronin, actin binding protein, 2B | 473 | 2.836 | 0.1095 | No |
| 6 | CDK6 | NA | CDK6 Entrez, Source | cyclin-dependent kinase 6 | 774 | 2.496 | 0.1170 | No |
| 7 | CFDP1 | NA | CFDP1 Entrez, Source | craniofacial development protein 1 | 912 | 2.381 | 0.1296 | No |
| 8 | ZIC4 | NA | ZIC4 Entrez, Source | Zic family member 4 | 1073 | 2.261 | 0.1405 | No |
| 9 | NRIP1 | NA | NRIP1 Entrez, Source | nuclear receptor interacting protein 1 | 1165 | 2.199 | 0.1535 | No |
| 10 | STX12 | NA | STX12 Entrez, Source | syntaxin 12 | 1193 | 2.181 | 0.1687 | No |
| 11 | SLC39A7 | NA | SLC39A7 Entrez, Source | solute carrier family 39 (zinc transporter), member 7 | 1772 | 1.839 | 0.1611 | No |
| 12 | TSPAN9 | NA | TSPAN9 Entrez, Source | tetraspanin 9 | 2055 | 1.695 | 0.1633 | No |
| 13 | NOS2 | NA | 2419 | 1.539 | 0.1613 | No | ||
| 14 | SWAP70 | NA | SWAP70 Entrez, Source | - | 2437 | 1.531 | 0.1721 | No |
| 15 | PTGDS | NA | PTGDS Entrez, Source | prostaglandin D2 synthase 21kDa (brain) | 2669 | 1.437 | 0.1742 | No |
| 16 | ST6GAL1 | NA | ST6GAL1 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 2690 | 1.425 | 0.1841 | No |
| 17 | GAP43 | NA | GAP43 Entrez, Source | growth associated protein 43 | 2743 | 1.403 | 0.1926 | No |
| 18 | NRG1 | NA | NRG1 Entrez, Source | neuregulin 1 | 3117 | 1.271 | 0.1883 | No |
| 19 | CPS1 | NA | CPS1 Entrez, Source | carbamoyl-phosphate synthetase 1, mitochondrial | 3203 | 1.238 | 0.1944 | No |
| 20 | EXOSC8 | NA | EXOSC8 Entrez, Source | exosome component 8 | 3415 | 1.179 | 0.1954 | No |
| 21 | SERINC1 | NA | SERINC1 Entrez, Source | serine incorporator 1 | 3639 | 1.102 | 0.1953 | No |
| 22 | ATXN2 | NA | ATXN2 Entrez, Source | ataxin 2 | 3818 | 1.044 | 0.1965 | No |
| 23 | FAM149B1 | NA | 3850 | 1.035 | 0.2031 | No | ||
| 24 | SNX19 | NA | SNX19 Entrez, Source | sorting nexin 19 | 4022 | 0.979 | 0.2041 | No |
| 25 | ZNF84 | NA | ZNF84 Entrez, Source | zinc finger protein 84 | 4076 | 0.965 | 0.2093 | No |
| 26 | DDN | NA | DDN Entrez, Source | dendrin | 4173 | 0.940 | 0.2127 | No |
| 27 | CLCN2 | NA | CLCN2 Entrez, Source | chloride channel 2 | 4410 | 0.881 | 0.2106 | No |
| 28 | HDLBP | NA | HDLBP Entrez, Source | high density lipoprotein binding protein (vigilin) | 4447 | 0.872 | 0.2157 | No |
| 29 | ALDH5A1 | NA | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 4715 | 0.806 | 0.2119 | No |
| 30 | TP53I3 | NA | TP53I3 Entrez, Source | tumor protein p53 inducible protein 3 | 5110 | 0.718 | 0.2027 | No |
| 31 | CCL25 | NA | CCL25 Entrez, Source | chemokine (C-C motif) ligand 25 | 5257 | 0.675 | 0.2024 | No |
| 32 | XPA | NA | XPA Entrez, Source | xeroderma pigmentosum, complementation group A | 5327 | 0.655 | 0.2047 | No |
| 33 | COL1A1 | NA | COL1A1 Entrez, Source | collagen, type I, alpha 1 | 5621 | 0.594 | 0.1983 | No |
| 34 | PHLDA2 | NA | PHLDA2 Entrez, Source | pleckstrin homology-like domain, family A, member 2 | 5724 | 0.575 | 0.1988 | No |
| 35 | REST | NA | REST Entrez, Source | RE1-silencing transcription factor | 5934 | 0.540 | 0.1952 | No |
| 36 | C7orf54 | NA | 6045 | 0.519 | 0.1950 | No | ||
| 37 | KBTBD11 | NA | KBTBD11 Entrez, Source | kelch repeat and BTB (POZ) domain containing 11 | 6292 | 0.482 | 0.1895 | No |
| 38 | CETN2 | NA | CETN2 Entrez, Source | centrin, EF-hand protein, 2 | 6441 | 0.459 | 0.1875 | No |
| 39 | ACP5 | NA | ACP5 Entrez, Source | acid phosphatase 5, tartrate resistant | 6502 | 0.453 | 0.1886 | No |
| 40 | CDC14B | NA | CDC14B Entrez, Source | CDC14 cell division cycle 14 homolog B (S. cerevisiae) | 6594 | 0.441 | 0.1886 | No |
| 41 | LEPROT | NA | LEPROT Entrez, Source | leptin receptor overlapping transcript | 6615 | 0.439 | 0.1911 | No |
| 42 | GNMT | NA | GNMT Entrez, Source | glycine N-methyltransferase | 6878 | 0.410 | 0.1845 | No |
| 43 | MTHFR | NA | MTHFR Entrez, Source | 5,10-methylenetetrahydrofolate reductase (NADPH) | 6944 | 0.403 | 0.1851 | No |
| 44 | MID1 | NA | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 7028 | 0.394 | 0.1850 | No |
| 45 | HYAL1 | NA | HYAL1 Entrez, Source | hyaluronoglucosaminidase 1 | 7390 | 0.355 | 0.1743 | No |
| 46 | VPS39 | NA | VPS39 Entrez, Source | vacuolar protein sorting 39 (yeast) | 7665 | 0.329 | 0.1667 | No |
| 47 | COL10A1 | NA | COL10A1 Entrez, Source | collagen, type X, alpha 1(Schmid metaphyseal chondrodysplasia) | 8007 | 0.300 | 0.1564 | No |
| 48 | C22orf31 | NA | C22ORF31 Entrez, Source | chromosome 22 open reading frame 31 | 8242 | 0.282 | 0.1499 | No |
| 49 | THOC2 | NA | THOC2 Entrez, Source | THO complex 2 | 8633 | 0.253 | 0.1374 | No |
| 50 | TAT | NA | TAT Entrez, Source | tyrosine aminotransferase | 8741 | 0.246 | 0.1353 | No |
| 51 | TCEAL2 | NA | TCEAL2 Entrez, Source | transcription elongation factor A (SII)-like 2 | 8803 | 0.241 | 0.1348 | No |
| 52 | ELP4 | NA | ELP4 Entrez, Source | elongation protein 4 homolog (S. cerevisiae) | 9276 | 0.215 | 0.1191 | No |
| 53 | PKN2 | NA | PKN2 Entrez, Source | protein kinase N2 | 9354 | 0.211 | 0.1178 | No |
| 54 | FCHSD2 | NA | FCHSD2 Entrez, Source | FCH and double SH3 domains 2 | 9702 | 0.193 | 0.1065 | No |
| 55 | C4orf6 | NA | C4ORF6 Entrez, Source | chromosome 4 open reading frame 6 | 9766 | 0.190 | 0.1056 | No |
| 56 | TAF12 | NA | TAF12 Entrez, Source | TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa | 10168 | 0.171 | 0.0921 | No |
| 57 | NUP160 | NA | NUP160 Entrez, Source | nucleoporin 160kDa | 10422 | 0.159 | 0.0840 | No |
| 58 | TNNT2 | NA | TNNT2 Entrez, Source | troponin T type 2 (cardiac) | 10748 | 0.144 | 0.0731 | No |
| 59 | FSTL4 | NA | FSTL4 Entrez, Source | follistatin-like 4 | 10888 | 0.138 | 0.0690 | No |
| 60 | KIR3DL2 | NA | KIR3DL2 Entrez, Source | killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 2 | 11258 | 0.120 | 0.0563 | No |
| 61 | MAML3 | NA | MAML3 Entrez, Source | mastermind-like 3 (Drosophila) | 11786 | 0.098 | 0.0377 | No |
| 62 | INHBA | NA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 11814 | 0.097 | 0.0374 | No |
| 63 | TBX5 | NA | TBX5 Entrez, Source | T-box 5 | 11889 | 0.094 | 0.0354 | No |
| 64 | TRAF5 | NA | TRAF5 Entrez, Source | TNF receptor-associated factor 5 | 12190 | 0.083 | 0.0250 | No |
| 65 | HCRTR1 | NA | HCRTR1 Entrez, Source | hypocretin (orexin) receptor 1 | 12423 | 0.074 | 0.0170 | No |
| 66 | NCKAP1L | NA | NCKAP1L Entrez, Source | NCK-associated protein 1-like | 12438 | 0.073 | 0.0170 | No |
| 67 | SRY | NA | SRY Entrez, Source | sex determining region Y | 12493 | 0.071 | 0.0155 | No |
| 68 | SERPINB2 | NA | SERPINB2 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 12515 | 0.070 | 0.0153 | No |
| 69 | AQP3 | NA | AQP3 Entrez, Source | aquaporin 3 (Gill blood group) | 12539 | 0.070 | 0.0150 | No |
| 70 | IL12RB1 | NA | IL12RB1 Entrez, Source | interleukin 12 receptor, beta 1 | 13025 | 0.052 | -0.0025 | No |
| 71 | NMBR | NA | NMBR Entrez, Source | neuromedin B receptor | 13117 | 0.048 | -0.0055 | No |
| 72 | NBAS | NA | 13250 | 0.043 | -0.0100 | No | ||
| 73 | GLRA2 | NA | GLRA2 Entrez, Source | glycine receptor, alpha 2 | 13641 | 0.028 | -0.0241 | No |
| 74 | NTNG1 | NA | NTNG1 Entrez, Source | netrin G1 | 14002 | 0.015 | -0.0373 | No |
| 75 | OPRK1 | NA | OPRK1 Entrez, Source | opioid receptor, kappa 1 | 14020 | 0.014 | -0.0378 | No |
| 76 | OCA2 | NA | OCA2 Entrez, Source | oculocutaneous albinism II (pink-eye dilution homolog, mouse) | 14175 | 0.009 | -0.0434 | No |
| 77 | NDUFS1 | NA | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 14276 | 0.005 | -0.0470 | No |
| 78 | MYBPC3 | NA | MYBPC3 Entrez, Source | myosin binding protein C, cardiac | 14931 | -0.018 | -0.0709 | No |
| 79 | KLK7 | NA | KLK7 Entrez, Source | kallikrein 7 (chymotryptic, stratum corneum) | 15820 | -0.053 | -0.1032 | No |
| 80 | PDIA3 | NA | PDIA3 Entrez, Source | protein disulfide isomerase family A, member 3 | 16486 | -0.080 | -0.1271 | No |
| 81 | CLEC2B | NA | CLEC2B Entrez, Source | C-type lectin domain family 2, member B | 16635 | -0.086 | -0.1319 | No |
| 82 | LRCH4 | NA | LRCH4 Entrez, Source | leucine-rich repeats and calponin homology (CH) domain containing 4 | 16978 | -0.100 | -0.1437 | No |
| 83 | SNRPD3 | NA | SNRPD3 Entrez, Source | small nuclear ribonucleoprotein D3 polypeptide 18kDa | 16999 | -0.101 | -0.1437 | No |
| 84 | GALNT3 | NA | GALNT3 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) | 17392 | -0.119 | -0.1572 | No |
| 85 | RALYL | NA | 17456 | -0.122 | -0.1586 | No | ||
| 86 | NDN | NA | NDN Entrez, Source | necdin homolog (mouse) | 18838 | -0.212 | -0.2078 | No |
| 87 | DSCR6 | NA | DSCR6 Entrez, Source | Down syndrome critical region gene 6 | 18851 | -0.214 | -0.2067 | No |
| 88 | LUZP2 | NA | LUZP2 Entrez, Source | leucine zipper protein 2 | 18856 | -0.214 | -0.2053 | No |
| 89 | DNAJC7 | NA | DNAJC7 Entrez, Source | DnaJ (Hsp40) homolog, subfamily C, member 7 | 19269 | -0.249 | -0.2186 | No |
| 90 | PAN2 | NA | 19507 | -0.271 | -0.2253 | No | ||
| 91 | OXCT1 | NA | OXCT1 Entrez, Source | 3-oxoacid CoA transferase 1 | 20005 | -0.321 | -0.2412 | No |
| 92 | MPI | NA | MPI Entrez, Source | mannose phosphate isomerase | 20515 | -0.382 | -0.2570 | No |
| 93 | MAGEA8 | NA | MAGEA8 Entrez, Source | melanoma antigen family A, 8 | 20635 | -0.397 | -0.2585 | No |
| 94 | ANXA10 | NA | ANXA10 Entrez, Source | annexin A10 | 20884 | -0.436 | -0.2644 | No |
| 95 | MRPS11 | NA | MRPS11 Entrez, Source | mitochondrial ribosomal protein S11 | 21198 | -0.480 | -0.2723 | No |
| 96 | MAOA | NA | MAOA Entrez, Source | monoamine oxidase A | 21228 | -0.486 | -0.2698 | No |
| 97 | AAGAB | NA | 21331 | -0.503 | -0.2698 | No | ||
| 98 | RBM3 | NA | RBM3 Entrez, Source | RNA binding motif (RNP1, RRM) protein 3 | 21476 | -0.529 | -0.2712 | No |
| 99 | TTC18 | NA | TTC18 Entrez, Source | tetratricopeptide repeat domain 18 | 21719 | -0.574 | -0.2758 | No |
| 100 | LMAN1 | NA | LMAN1 Entrez, Source | lectin, mannose-binding, 1 | 21774 | -0.586 | -0.2734 | No |
| 101 | SPAG5 | NA | SPAG5 Entrez, Source | sperm associated antigen 5 | 21827 | -0.597 | -0.2709 | No |
| 102 | FTSJD2 | NA | 22256 | -0.678 | -0.2816 | No | ||
| 103 | NEU3 | NA | NEU3 Entrez, Source | sialidase 3 (membrane sialidase) | 22517 | -0.733 | -0.2857 | No |
| 104 | EXOSC10 | NA | EXOSC10 Entrez, Source | exosome component 10 | 22527 | -0.735 | -0.2806 | No |
| 105 | LOC81691 | NA | LOC81691 Entrez, Source | - | 22650 | -0.765 | -0.2794 | No |
| 106 | KIFAP3 | NA | KIFAP3 Entrez, Source | kinesin-associated protein 3 | 22852 | -0.807 | -0.2808 | No |
| 107 | SNRNP40 | NA | 23089 | -0.856 | -0.2831 | No | ||
| 108 | ATXN7 | NA | ATXN7 Entrez, Source | ataxin 7 | 23413 | -0.929 | -0.2881 | Yes |
| 109 | CALML4 | NA | CALML4 Entrez, Source | calmodulin-like 4 | 23553 | -0.966 | -0.2861 | Yes |
| 110 | AZI1 | NA | AZI1 Entrez, Source | 5-azacytidine induced 1 | 23573 | -0.971 | -0.2795 | Yes |
| 111 | RTCD1 | NA | RTCD1 Entrez, Source | RNA terminal phosphate cyclase domain 1 | 23588 | -0.977 | -0.2728 | Yes |
| 112 | RRBP1 | NA | RRBP1 Entrez, Source | ribosome binding protein 1 homolog 180kDa (dog) | 23695 | -1.007 | -0.2692 | Yes |
| 113 | ATF6 | NA | ATF6 Entrez, Source | activating transcription factor 6 | 24198 | -1.147 | -0.2792 | Yes |
| 114 | MYO1E | NA | MYO1E Entrez, Source | myosin IE | 24442 | -1.224 | -0.2790 | Yes |
| 115 | LRPAP1 | NA | LRPAP1 Entrez, Source | low density lipoprotein receptor-related protein associated protein 1 | 24477 | -1.235 | -0.2711 | Yes |
| 116 | EXOSC7 | NA | EXOSC7 Entrez, Source | exosome component 7 | 24684 | -1.303 | -0.2690 | Yes |
| 117 | RPIA | NA | RPIA Entrez, Source | ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) | 24742 | -1.324 | -0.2613 | Yes |
| 118 | ANXA7 | NA | ANXA7 Entrez, Source | annexin A7 | 24823 | -1.351 | -0.2542 | Yes |
| 119 | MVK | NA | MVK Entrez, Source | mevalonate kinase (mevalonic aciduria) | 24824 | -1.351 | -0.2442 | Yes |
| 120 | ACOT11 | NA | ACOT11 Entrez, Source | acyl-CoA thioesterase 11 | 25249 | -1.521 | -0.2485 | Yes |
| 121 | CREG1 | NA | CREG1 Entrez, Source | cellular repressor of E1A-stimulated genes 1 | 25519 | -1.649 | -0.2461 | Yes |
| 122 | GLRX | NA | GLRX Entrez, Source | glutaredoxin (thioltransferase) | 25564 | -1.671 | -0.2353 | Yes |
| 123 | CHEK2 | NA | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 25612 | -1.694 | -0.2245 | Yes |
| 124 | ADCY7 | NA | ADCY7 Entrez, Source | adenylate cyclase 7 | 25733 | -1.754 | -0.2159 | Yes |
| 125 | C17orf75 | NA | C17ORF75 Entrez, Source | chromosome 17 open reading frame 75 | 26024 | -1.935 | -0.2122 | Yes |
| 126 | HSPA4L | NA | HSPA4L Entrez, Source | heat shock 70kDa protein 4-like | 26273 | -2.108 | -0.2057 | Yes |
| 127 | ANXA11 | NA | ANXA11 Entrez, Source | annexin A11 | 26366 | -2.184 | -0.1928 | Yes |
| 128 | FLI1 | NA | FLI1 Entrez, Source | Friend leukemia virus integration 1 | 26467 | -2.267 | -0.1797 | Yes |
| 129 | ZNF185 | NA | ZNF185 Entrez, Source | zinc finger protein 185 (LIM domain) | 26572 | -2.357 | -0.1660 | Yes |
| 130 | HEBP2 | NA | HEBP2 Entrez, Source | heme binding protein 2 | 26586 | -2.372 | -0.1489 | Yes |
| 131 | TAF1B | NA | TAF1B Entrez, Source | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa | 26594 | -2.379 | -0.1315 | Yes |
| 132 | PSMB10 | NA | PSMB10 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 10 | 26623 | -2.409 | -0.1146 | Yes |
| 133 | RARS | NA | RARS Entrez, Source | arginyl-tRNA synthetase | 26640 | -2.433 | -0.0972 | Yes |
| 134 | PPIC | NA | PPIC Entrez, Source | peptidylprolyl isomerase C (cyclophilin C) | 26687 | -2.486 | -0.0804 | Yes |
| 135 | GTF2H5 | NA | GTF2H5 Entrez, Source | general transcription factor IIH, polypeptide 5 | 26979 | -2.976 | -0.0690 | Yes |
| 136 | LAS1L | NA | LAS1L Entrez, Source | LAS1-like (S. cerevisiae) | 27018 | -3.083 | -0.0475 | Yes |
| 137 | PAX2 | NA | PAX2 Entrez, Source | paired box gene 2 | 27125 | -3.450 | -0.0258 | Yes |
| 138 | NEK9 | NA | NEK9 Entrez, Source | NIMA (never in mitosis gene a)- related kinase 9 | 27289 | -4.485 | 0.0015 | Yes |