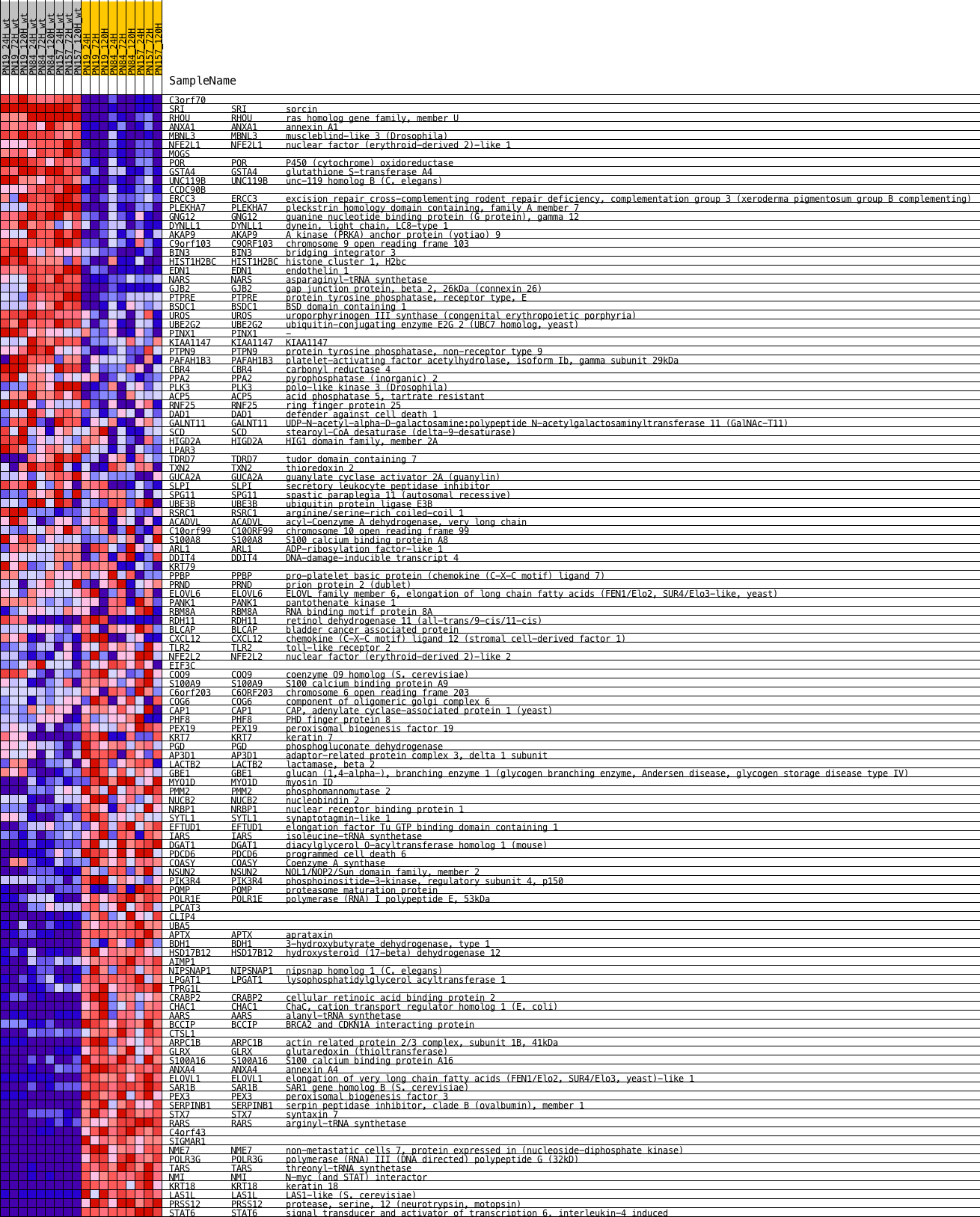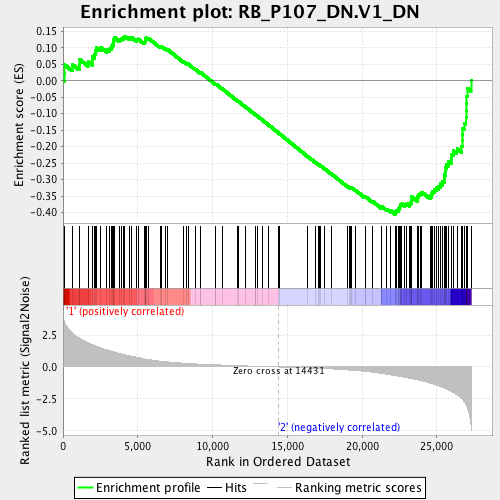
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | RB_P107_DN.V1_DN |
| Enrichment Score (ES) | -0.4059992 |
| Normalized Enrichment Score (NES) | -1.4781135 |
| Nominal p-value | 0.016 |
| FDR q-value | 0.19717456 |
| FWER p-Value | 0.311 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | C3orf70 | NA | 90 | 3.509 | 0.0228 | No | ||
| 2 | SRI | NA | SRI Entrez, Source | sorcin | 99 | 3.489 | 0.0484 | No |
| 3 | RHOU | NA | RHOU Entrez, Source | ras homolog gene family, member U | 609 | 2.667 | 0.0495 | No |
| 4 | ANXA1 | NA | ANXA1 Entrez, Source | annexin A1 | 1067 | 2.267 | 0.0495 | No |
| 5 | MBNL3 | NA | MBNL3 Entrez, Source | muscleblind-like 3 (Drosophila) | 1120 | 2.234 | 0.0642 | No |
| 6 | NFE2L1 | NA | NFE2L1 Entrez, Source | nuclear factor (erythroid-derived 2)-like 1 | 1666 | 1.889 | 0.0582 | No |
| 7 | MOGS | NA | 1960 | 1.745 | 0.0604 | No | ||
| 8 | POR | NA | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 1971 | 1.740 | 0.0729 | No |
| 9 | GSTA4 | NA | GSTA4 Entrez, Source | glutathione S-transferase A4 | 2105 | 1.675 | 0.0805 | No |
| 10 | UNC119B | NA | UNC119B Entrez, Source | unc-119 homolog B (C. elegans) | 2168 | 1.646 | 0.0904 | No |
| 11 | CCDC90B | NA | 2207 | 1.630 | 0.1011 | No | ||
| 12 | ERCC3 | NA | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 2486 | 1.511 | 0.1021 | No |
| 13 | PLEKHA7 | NA | PLEKHA7 Entrez, Source | pleckstrin homology domain containing, family A member 7 | 2932 | 1.337 | 0.0957 | No |
| 14 | GNG12 | NA | GNG12 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 12 | 3133 | 1.263 | 0.0977 | No |
| 15 | DYNLL1 | NA | DYNLL1 Entrez, Source | dynein, light chain, LC8-type 1 | 3244 | 1.227 | 0.1028 | No |
| 16 | AKAP9 | NA | AKAP9 Entrez, Source | A kinase (PRKA) anchor protein (yotiao) 9 | 3325 | 1.207 | 0.1088 | No |
| 17 | C9orf103 | NA | C9ORF103 Entrez, Source | chromosome 9 open reading frame 103 | 3367 | 1.195 | 0.1162 | No |
| 18 | BIN3 | NA | BIN3 Entrez, Source | bridging integrator 3 | 3396 | 1.184 | 0.1239 | No |
| 19 | HIST1H2BC | NA | HIST1H2BC Entrez, Source | histone cluster 1, H2bc | 3447 | 1.169 | 0.1308 | No |
| 20 | EDN1 | NA | EDN1 Entrez, Source | endothelin 1 | 3804 | 1.049 | 0.1255 | No |
| 21 | NARS | NA | NARS Entrez, Source | asparaginyl-tRNA synthetase | 3934 | 1.007 | 0.1282 | No |
| 22 | GJB2 | NA | GJB2 Entrez, Source | gap junction protein, beta 2, 26kDa (connexin 26) | 4039 | 0.976 | 0.1317 | No |
| 23 | PTPRE | NA | PTPRE Entrez, Source | protein tyrosine phosphatase, receptor type, E | 4129 | 0.951 | 0.1354 | No |
| 24 | BSDC1 | NA | BSDC1 Entrez, Source | BSD domain containing 1 | 4434 | 0.875 | 0.1308 | No |
| 25 | UROS | NA | UROS Entrez, Source | uroporphyrinogen III synthase (congenital erythropoietic porphyria) | 4567 | 0.847 | 0.1322 | No |
| 26 | UBE2G2 | NA | UBE2G2 Entrez, Source | ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) | 4907 | 0.765 | 0.1254 | No |
| 27 | PINX1 | NA | PINX1 Entrez, Source | - | 5022 | 0.739 | 0.1267 | No |
| 28 | KIAA1147 | NA | KIAA1147 Entrez, Source | KIAA1147 | 5417 | 0.635 | 0.1170 | No |
| 29 | PTPN9 | NA | PTPN9 Entrez, Source | protein tyrosine phosphatase, non-receptor type 9 | 5482 | 0.622 | 0.1192 | No |
| 30 | PAFAH1B3 | NA | PAFAH1B3 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | 5493 | 0.619 | 0.1235 | No |
| 31 | CBR4 | NA | CBR4 Entrez, Source | carbonyl reductase 4 | 5511 | 0.615 | 0.1274 | No |
| 32 | PPA2 | NA | PPA2 Entrez, Source | pyrophosphatase (inorganic) 2 | 5564 | 0.603 | 0.1300 | No |
| 33 | PLK3 | NA | PLK3 Entrez, Source | polo-like kinase 3 (Drosophila) | 5736 | 0.573 | 0.1279 | No |
| 34 | ACP5 | NA | ACP5 Entrez, Source | acid phosphatase 5, tartrate resistant | 6502 | 0.453 | 0.1032 | No |
| 35 | RNF25 | NA | RNF25 Entrez, Source | ring finger protein 25 | 6574 | 0.444 | 0.1039 | No |
| 36 | DAD1 | NA | DAD1 Entrez, Source | defender against cell death 1 | 6817 | 0.416 | 0.0981 | No |
| 37 | GALNT11 | NA | GALNT11 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) | 6958 | 0.401 | 0.0959 | No |
| 38 | SCD | NA | SCD Entrez, Source | stearoyl-CoA desaturase (delta-9-desaturase) | 8046 | 0.296 | 0.0581 | No |
| 39 | HIGD2A | NA | HIGD2A Entrez, Source | HIG1 domain family, member 2A | 8253 | 0.281 | 0.0527 | No |
| 40 | LPAR3 | NA | 8355 | 0.273 | 0.0510 | No | ||
| 41 | TDRD7 | NA | TDRD7 Entrez, Source | tudor domain containing 7 | 8833 | 0.240 | 0.0352 | No |
| 42 | TXN2 | NA | TXN2 Entrez, Source | thioredoxin 2 | 9165 | 0.221 | 0.0247 | No |
| 43 | GUCA2A | NA | GUCA2A Entrez, Source | guanylate cyclase activator 2A (guanylin) | 9185 | 0.220 | 0.0256 | No |
| 44 | SLPI | NA | SLPI Entrez, Source | secretory leukocyte peptidase inhibitor | 10226 | 0.168 | -0.0114 | No |
| 45 | SPG11 | NA | SPG11 Entrez, Source | spastic paraplegia 11 (autosomal recessive) | 10227 | 0.168 | -0.0101 | No |
| 46 | UBE3B | NA | UBE3B Entrez, Source | ubiquitin protein ligase E3B | 10637 | 0.150 | -0.0240 | No |
| 47 | RSRC1 | NA | RSRC1 Entrez, Source | arginine/serine-rich coiled-coil 1 | 11637 | 0.105 | -0.0600 | No |
| 48 | ACADVL | NA | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 11725 | 0.101 | -0.0624 | No |
| 49 | C10orf99 | NA | C10ORF99 Entrez, Source | chromosome 10 open reading frame 99 | 12203 | 0.082 | -0.0793 | No |
| 50 | S100A8 | NA | S100A8 Entrez, Source | S100 calcium binding protein A8 | 12854 | 0.059 | -0.1028 | No |
| 51 | ARL1 | NA | ARL1 Entrez, Source | ADP-ribosylation factor-like 1 | 12881 | 0.058 | -0.1033 | No |
| 52 | DDIT4 | NA | DDIT4 Entrez, Source | DNA-damage-inducible transcript 4 | 13010 | 0.053 | -0.1076 | No |
| 53 | KRT79 | NA | 13365 | 0.039 | -0.1204 | No | ||
| 54 | PPBP | NA | PPBP Entrez, Source | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | 13740 | 0.024 | -0.1339 | No |
| 55 | PRND | NA | PRND Entrez, Source | prion protein 2 (dublet) | 14440 | -0.000 | -0.1596 | No |
| 56 | ELOVL6 | NA | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 14477 | -0.002 | -0.1609 | No |
| 57 | PANK1 | NA | PANK1 Entrez, Source | pantothenate kinase 1 | 16336 | -0.075 | -0.2287 | No |
| 58 | RBM8A | NA | RBM8A Entrez, Source | RNA binding motif protein 8A | 16892 | -0.097 | -0.2484 | No |
| 59 | RDH11 | NA | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 17069 | -0.104 | -0.2541 | No |
| 60 | BLCAP | NA | BLCAP Entrez, Source | bladder cancer associated protein | 17124 | -0.107 | -0.2553 | No |
| 61 | CXCL12 | NA | CXCL12 Entrez, Source | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 17216 | -0.111 | -0.2578 | No |
| 62 | TLR2 | NA | TLR2 Entrez, Source | toll-like receptor 2 | 17512 | -0.125 | -0.2677 | No |
| 63 | NFE2L2 | NA | NFE2L2 Entrez, Source | nuclear factor (erythroid-derived 2)-like 2 | 17949 | -0.150 | -0.2826 | No |
| 64 | EIF3C | NA | 19044 | -0.231 | -0.3211 | No | ||
| 65 | COQ9 | NA | COQ9 Entrez, Source | coenzyme Q9 homolog (S. cerevisiae) | 19172 | -0.241 | -0.3240 | No |
| 66 | S100A9 | NA | S100A9 Entrez, Source | S100 calcium binding protein A9 | 19218 | -0.246 | -0.3238 | No |
| 67 | C6orf203 | NA | C6ORF203 Entrez, Source | chromosome 6 open reading frame 203 | 19268 | -0.249 | -0.3238 | No |
| 68 | COG6 | NA | COG6 Entrez, Source | component of oligomeric golgi complex 6 | 19566 | -0.277 | -0.3326 | No |
| 69 | CAP1 | NA | CAP1 Entrez, Source | CAP, adenylate cyclase-associated protein 1 (yeast) | 20201 | -0.341 | -0.3534 | No |
| 70 | PHF8 | NA | PHF8 Entrez, Source | PHD finger protein 8 | 20207 | -0.342 | -0.3510 | No |
| 71 | PEX19 | NA | PEX19 Entrez, Source | peroxisomal biogenesis factor 19 | 20709 | -0.407 | -0.3664 | No |
| 72 | KRT7 | NA | KRT7 Entrez, Source | keratin 7 | 21305 | -0.497 | -0.3846 | No |
| 73 | PGD | NA | PGD Entrez, Source | phosphogluconate dehydrogenase | 21318 | -0.500 | -0.3813 | No |
| 74 | AP3D1 | NA | AP3D1 Entrez, Source | adaptor-related protein complex 3, delta 1 subunit | 21651 | -0.561 | -0.3894 | No |
| 75 | LACTB2 | NA | LACTB2 Entrez, Source | lactamase, beta 2 | 21922 | -0.612 | -0.3948 | No |
| 76 | GBE1 | NA | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 (glycogen branching enzyme, Andersen disease, glycogen storage disease type IV) | 22229 | -0.671 | -0.4010 | Yes |
| 77 | MYO1D | NA | MYO1D Entrez, Source | myosin ID | 22262 | -0.680 | -0.3971 | Yes |
| 78 | PMM2 | NA | PMM2 Entrez, Source | phosphomannomutase 2 | 22327 | -0.692 | -0.3944 | Yes |
| 79 | NUCB2 | NA | NUCB2 Entrez, Source | nucleobindin 2 | 22419 | -0.711 | -0.3924 | Yes |
| 80 | NRBP1 | NA | NRBP1 Entrez, Source | nuclear receptor binding protein 1 | 22438 | -0.715 | -0.3878 | Yes |
| 81 | SYTL1 | NA | SYTL1 Entrez, Source | synaptotagmin-like 1 | 22529 | -0.736 | -0.3856 | Yes |
| 82 | EFTUD1 | NA | EFTUD1 Entrez, Source | elongation factor Tu GTP binding domain containing 1 | 22541 | -0.740 | -0.3805 | Yes |
| 83 | IARS | NA | IARS Entrez, Source | isoleucine-tRNA synthetase | 22588 | -0.750 | -0.3767 | Yes |
| 84 | DGAT1 | NA | DGAT1 Entrez, Source | diacylglycerol O-acyltransferase homolog 1 (mouse) | 22671 | -0.768 | -0.3740 | Yes |
| 85 | PDCD6 | NA | PDCD6 Entrez, Source | programmed cell death 6 | 22862 | -0.809 | -0.3749 | Yes |
| 86 | COASY | NA | COASY Entrez, Source | Coenzyme A synthase | 23004 | -0.840 | -0.3739 | Yes |
| 87 | NSUN2 | NA | NSUN2 Entrez, Source | NOL1/NOP2/Sun domain family, member 2 | 23184 | -0.874 | -0.3740 | Yes |
| 88 | PIK3R4 | NA | PIK3R4 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 4, p150 | 23271 | -0.896 | -0.3705 | Yes |
| 89 | POMP | NA | POMP Entrez, Source | proteasome maturation protein | 23306 | -0.903 | -0.3650 | Yes |
| 90 | POLR1E | NA | POLR1E Entrez, Source | polymerase (RNA) I polypeptide E, 53kDa | 23322 | -0.906 | -0.3588 | Yes |
| 91 | LPCAT3 | NA | 23324 | -0.906 | -0.3521 | Yes | ||
| 92 | CLIP4 | NA | 23716 | -1.012 | -0.3590 | Yes | ||
| 93 | UBA5 | NA | 23726 | -1.013 | -0.3518 | Yes | ||
| 94 | APTX | NA | APTX Entrez, Source | aprataxin | 23794 | -1.034 | -0.3466 | Yes |
| 95 | BDH1 | NA | BDH1 Entrez, Source | 3-hydroxybutyrate dehydrogenase, type 1 | 23879 | -1.059 | -0.3418 | Yes |
| 96 | HSD17B12 | NA | HSD17B12 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 12 | 23989 | -1.086 | -0.3377 | Yes |
| 97 | AIMP1 | NA | 24553 | -1.262 | -0.3491 | Yes | ||
| 98 | NIPSNAP1 | NA | NIPSNAP1 Entrez, Source | nipsnap homolog 1 (C. elegans) | 24666 | -1.297 | -0.3436 | Yes |
| 99 | LPGAT1 | NA | LPGAT1 Entrez, Source | lysophosphatidylglycerol acyltransferase 1 | 24698 | -1.308 | -0.3350 | Yes |
| 100 | TPRG1L | NA | 24852 | -1.361 | -0.3305 | Yes | ||
| 101 | CRABP2 | NA | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 24956 | -1.400 | -0.3239 | Yes |
| 102 | CHAC1 | NA | CHAC1 Entrez, Source | ChaC, cation transport regulator homolog 1 (E. coli) | 25142 | -1.481 | -0.3197 | Yes |
| 103 | AARS | NA | AARS Entrez, Source | alanyl-tRNA synthetase | 25263 | -1.527 | -0.3128 | Yes |
| 104 | BCCIP | NA | BCCIP Entrez, Source | BRCA2 and CDKN1A interacting protein | 25383 | -1.584 | -0.3054 | Yes |
| 105 | CTSL1 | NA | 25492 | -1.639 | -0.2972 | Yes | ||
| 106 | ARPC1B | NA | ARPC1B Entrez, Source | actin related protein 2/3 complex, subunit 1B, 41kDa | 25503 | -1.643 | -0.2853 | Yes |
| 107 | GLRX | NA | GLRX Entrez, Source | glutaredoxin (thioltransferase) | 25564 | -1.671 | -0.2751 | Yes |
| 108 | S100A16 | NA | S100A16 Entrez, Source | S100 calcium binding protein A16 | 25580 | -1.679 | -0.2632 | Yes |
| 109 | ANXA4 | NA | ANXA4 Entrez, Source | annexin A4 | 25649 | -1.710 | -0.2530 | Yes |
| 110 | ELOVL1 | NA | ELOVL1 Entrez, Source | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 | 25811 | -1.803 | -0.2456 | Yes |
| 111 | SAR1B | NA | SAR1B Entrez, Source | SAR1 gene homolog B (S. cerevisiae) | 25961 | -1.892 | -0.2370 | Yes |
| 112 | PEX3 | NA | PEX3 Entrez, Source | peroxisomal biogenesis factor 3 | 26010 | -1.926 | -0.2244 | Yes |
| 113 | SERPINB1 | NA | SERPINB1 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 1 | 26100 | -1.980 | -0.2130 | Yes |
| 114 | STX7 | NA | STX7 Entrez, Source | syntaxin 7 | 26354 | -2.176 | -0.2061 | Yes |
| 115 | RARS | NA | RARS Entrez, Source | arginyl-tRNA synthetase | 26640 | -2.433 | -0.1985 | Yes |
| 116 | C4orf43 | NA | 26710 | -2.520 | -0.1824 | Yes | ||
| 117 | SIGMAR1 | NA | 26722 | -2.531 | -0.1640 | Yes | ||
| 118 | NME7 | NA | NME7 Entrez, Source | non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) | 26746 | -2.557 | -0.1458 | Yes |
| 119 | POLR3G | NA | POLR3G Entrez, Source | polymerase (RNA) III (DNA directed) polypeptide G (32kD) | 26828 | -2.699 | -0.1288 | Yes |
| 120 | TARS | NA | TARS Entrez, Source | threonyl-tRNA synthetase | 26956 | -2.925 | -0.1117 | Yes |
| 121 | NMI | NA | NMI Entrez, Source | N-myc (and STAT) interactor | 26969 | -2.946 | -0.0903 | Yes |
| 122 | KRT18 | NA | KRT18 Entrez, Source | keratin 18 | 27009 | -3.061 | -0.0690 | Yes |
| 123 | LAS1L | NA | LAS1L Entrez, Source | LAS1-like (S. cerevisiae) | 27018 | -3.083 | -0.0464 | Yes |
| 124 | PRSS12 | NA | PRSS12 Entrez, Source | protease, serine, 12 (neurotrypsin, motopsin) | 27076 | -3.264 | -0.0242 | Yes |
| 125 | STAT6 | NA | STAT6 Entrez, Source | signal transducer and activator of transcription 6, interleukin-4 induced | 27292 | -4.512 | 0.0014 | Yes |