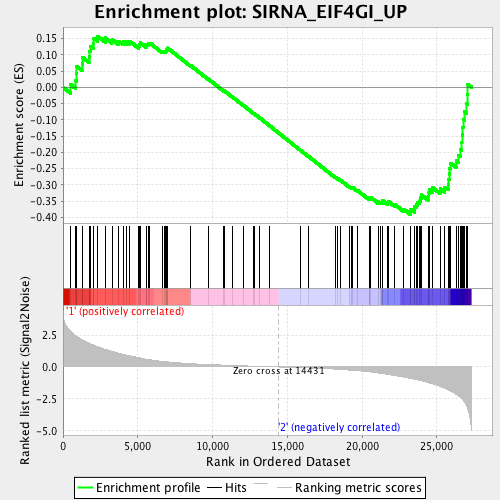
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | SIRNA_EIF4GI_UP |
| Enrichment Score (ES) | -0.39076936 |
| Normalized Enrichment Score (NES) | -1.3986592 |
| Nominal p-value | 0.025477707 |
| FDR q-value | 0.2256265 |
| FWER p-Value | 0.524 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ASAH1 | NA | ASAH1 Entrez, Source | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 501 | 2.792 | 0.0089 | No |
| 2 | TXNIP | NA | TXNIP Entrez, Source | thioredoxin interacting protein | 806 | 2.466 | 0.0218 | No |
| 3 | C7orf10 | NA | C7ORF10 Entrez, Source | chromosome 7 open reading frame 10 | 876 | 2.409 | 0.0428 | No |
| 4 | PSAP | NA | PSAP Entrez, Source | prosaposin (variant Gaucher disease and variant metachromatic leukodystrophy) | 903 | 2.388 | 0.0651 | No |
| 5 | PMPCB | NA | PMPCB Entrez, Source | peptidase (mitochondrial processing) beta | 1266 | 2.126 | 0.0726 | No |
| 6 | TCF25 | NA | TCF25 Entrez, Source | transcription factor 25 (basic helix-loop-helix) | 1298 | 2.102 | 0.0920 | No |
| 7 | CA2 | NA | CA2 Entrez, Source | carbonic anhydrase II | 1735 | 1.854 | 0.0941 | No |
| 8 | DAG1 | NA | DAG1 Entrez, Source | dystroglycan 1 (dystrophin-associated glycoprotein 1) | 1782 | 1.834 | 0.1103 | No |
| 9 | PTPN13 | NA | PTPN13 Entrez, Source | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | 1829 | 1.809 | 0.1262 | No |
| 10 | BTG2 | NA | BTG2 Entrez, Source | BTG family, member 2 | 2049 | 1.697 | 0.1348 | No |
| 11 | ATP5A1 | NA | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 2058 | 1.693 | 0.1510 | No |
| 12 | PAPSS1 | NA | PAPSS1 Entrez, Source | 3'-phosphoadenosine 5'-phosphosulfate synthase 1 | 2309 | 1.591 | 0.1574 | No |
| 13 | TCTN1 | NA | 2814 | 1.381 | 0.1523 | No | ||
| 14 | MAGEF1 | NA | MAGEF1 Entrez, Source | melanoma antigen family F, 1 | 3271 | 1.221 | 0.1475 | No |
| 15 | IGF2R | NA | IGF2R Entrez, Source | insulin-like growth factor 2 receptor | 3724 | 1.075 | 0.1414 | No |
| 16 | UBE2E1 | NA | UBE2E1 Entrez, Source | ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast) | 4020 | 0.979 | 0.1402 | No |
| 17 | ATF3 | NA | ATF3 Entrez, Source | activating transcription factor 3 | 4220 | 0.927 | 0.1419 | No |
| 18 | KIF2A | NA | KIF2A Entrez, Source | kinesin heavy chain member 2A | 4466 | 0.869 | 0.1414 | No |
| 19 | EID1 | NA | EID1 Entrez, Source | EP300 interacting inhibitor of differentiation 1 | 5062 | 0.731 | 0.1267 | No |
| 20 | CTSH | NA | CTSH Entrez, Source | cathepsin H | 5119 | 0.715 | 0.1316 | No |
| 21 | ZFP36L2 | NA | ZFP36L2 Entrez, Source | zinc finger protein 36, C3H type-like 2 | 5144 | 0.709 | 0.1376 | No |
| 22 | HSPB1 | NA | HSPB1 Entrez, Source | heat shock 27kDa protein 1 | 5569 | 0.602 | 0.1280 | No |
| 23 | PYGL | NA | PYGL Entrez, Source | phosphorylase, glycogen; liver (Hers disease, glycogen storage disease type VI) | 5608 | 0.595 | 0.1324 | No |
| 24 | INPP5D | NA | INPP5D Entrez, Source | inositol polyphosphate-5-phosphatase, 145kDa | 5714 | 0.576 | 0.1341 | No |
| 25 | NQO1 | NA | NQO1 Entrez, Source | NAD(P)H dehydrogenase, quinone 1 | 5811 | 0.558 | 0.1361 | No |
| 26 | FBXO46 | NA | FBXO46 Entrez, Source | F-box protein 46 | 6626 | 0.437 | 0.1105 | No |
| 27 | TRAF3IP2 | NA | TRAF3IP2 Entrez, Source | TRAF3 interacting protein 2 | 6749 | 0.424 | 0.1101 | No |
| 28 | D4S234E | NA | D4S234E Entrez, Source | - | 6856 | 0.412 | 0.1102 | No |
| 29 | MPV17 | NA | MPV17 Entrez, Source | MpV17 mitochondrial inner membrane protein | 6887 | 0.409 | 0.1131 | No |
| 30 | ATP1A1 | NA | ATP1A1 Entrez, Source | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 6929 | 0.404 | 0.1156 | No |
| 31 | HTRA2 | NA | HTRA2 Entrez, Source | HtrA serine peptidase 2 | 6996 | 0.397 | 0.1170 | No |
| 32 | HADHB | NA | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 6998 | 0.397 | 0.1209 | No |
| 33 | CEBPD | NA | CEBPD Entrez, Source | CCAAT/enhancer binding protein (C/EBP), delta | 8552 | 0.259 | 0.0664 | No |
| 34 | ASS1 | NA | ASS1 Entrez, Source | argininosuccinate synthetase 1 | 9695 | 0.193 | 0.0263 | No |
| 35 | NONO | NA | NONO Entrez, Source | non-POU domain containing, octamer-binding | 10725 | 0.145 | -0.0100 | No |
| 36 | FXYD3 | NA | FXYD3 Entrez, Source | FXYD domain containing ion transport regulator 3 | 10766 | 0.143 | -0.0101 | No |
| 37 | SPRR1A | NA | SPRR1A Entrez, Source | small proline-rich protein 1A | 11314 | 0.117 | -0.0290 | No |
| 38 | CAMLG | NA | CAMLG Entrez, Source | calcium modulating ligand | 12044 | 0.089 | -0.0549 | No |
| 39 | FCAR | NA | FCAR Entrez, Source | Fc fragment of IgA, receptor for | 12725 | 0.063 | -0.0793 | No |
| 40 | XPC | NA | XPC Entrez, Source | xeroderma pigmentosum, complementation group C | 12806 | 0.060 | -0.0816 | No |
| 41 | SPRR1B | NA | SPRR1B Entrez, Source | small proline-rich protein 1B (cornifin) | 13126 | 0.048 | -0.0929 | No |
| 42 | QARS | NA | QARS Entrez, Source | glutaminyl-tRNA synthetase | 13829 | 0.021 | -0.1184 | No |
| 43 | SETDB1 | NA | SETDB1 Entrez, Source | SET domain, bifurcated 1 | 15884 | -0.055 | -0.1933 | No |
| 44 | MATR3 | NA | MATR3 Entrez, Source | matrin 3 | 16439 | -0.079 | -0.2129 | No |
| 45 | WISP2 | NA | WISP2 Entrez, Source | WNT1 inducible signaling pathway protein 2 | 18222 | -0.167 | -0.2767 | No |
| 46 | GPR15 | NA | GPR15 Entrez, Source | G protein-coupled receptor 15 | 18372 | -0.176 | -0.2804 | No |
| 47 | SAMM50 | NA | SAMM50 Entrez, Source | sorting and assembly machinery component 50 homolog (S. cerevisiae) | 18572 | -0.190 | -0.2859 | No |
| 48 | SLCO1A2 | NA | SLCO1A2 Entrez, Source | solute carrier organic anion transporter family, member 1A2 | 19188 | -0.243 | -0.3061 | No |
| 49 | SPG20 | NA | SPG20 Entrez, Source | spastic paraplegia 20, spartin (Troyer syndrome) | 19288 | -0.250 | -0.3073 | No |
| 50 | SAA1 | NA | SAA1 Entrez, Source | serum amyloid A1 | 19376 | -0.258 | -0.3080 | No |
| 51 | DDX54 | NA | DDX54 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | 19681 | -0.287 | -0.3163 | No |
| 52 | DDX50 | NA | DDX50 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 | 20495 | -0.380 | -0.3425 | No |
| 53 | GIN1 | NA | 20519 | -0.382 | -0.3396 | No | ||
| 54 | ST13 | NA | ST13 Entrez, Source | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 20565 | -0.389 | -0.3374 | No |
| 55 | DTX4 | NA | DTX4 Entrez, Source | deltex 4 homolog (Drosophila) | 21079 | -0.464 | -0.3517 | No |
| 56 | S100A14 | NA | S100A14 Entrez, Source | S100 calcium binding protein A14 | 21230 | -0.486 | -0.3525 | No |
| 57 | FRAT2 | NA | FRAT2 Entrez, Source | frequently rearranged in advanced T-cell lymphomas 2 | 21360 | -0.508 | -0.3523 | No |
| 58 | GSS | NA | GSS Entrez, Source | glutathione synthetase | 21373 | -0.510 | -0.3477 | No |
| 59 | NCAPD2 | NA | NCAPD2 Entrez, Source | non-SMC condensin I complex, subunit D2 | 21700 | -0.570 | -0.3541 | No |
| 60 | GLUD1 | NA | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 21742 | -0.579 | -0.3500 | No |
| 61 | SLC25A3 | NA | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 22165 | -0.659 | -0.3590 | No |
| 62 | EIF3K | NA | 22791 | -0.793 | -0.3742 | No | ||
| 63 | SLC25A5 | NA | SLC25A5 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 23242 | -0.890 | -0.3821 | Yes |
| 64 | DRG1 | NA | DRG1 Entrez, Source | developmentally regulated GTP binding protein 1 | 23270 | -0.896 | -0.3743 | Yes |
| 65 | UQCRC2 | NA | UQCRC2 Entrez, Source | ubiquinol-cytochrome c reductase core protein II | 23520 | -0.957 | -0.3741 | Yes |
| 66 | CPNE1 | NA | CPNE1 Entrez, Source | copine I | 23536 | -0.960 | -0.3653 | Yes |
| 67 | CSDE1 | NA | CSDE1 Entrez, Source | cold shock domain containing E1, RNA-binding | 23635 | -0.988 | -0.3593 | Yes |
| 68 | RMND1 | NA | 23734 | -1.016 | -0.3529 | Yes | ||
| 69 | LETMD1 | NA | LETMD1 Entrez, Source | LETM1 domain containing 1 | 23876 | -1.059 | -0.3478 | Yes |
| 70 | PSME1 | NA | PSME1 Entrez, Source | proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) | 23933 | -1.073 | -0.3394 | Yes |
| 71 | THADA | NA | THADA Entrez, Source | thyroid adenoma associated | 23987 | -1.086 | -0.3307 | Yes |
| 72 | ZBED1 | NA | ZBED1 Entrez, Source | zinc finger, BED-type containing 1 | 24457 | -1.229 | -0.3359 | Yes |
| 73 | LRPAP1 | NA | LRPAP1 Entrez, Source | low density lipoprotein receptor-related protein associated protein 1 | 24477 | -1.235 | -0.3246 | Yes |
| 74 | ESYT1 | NA | 24537 | -1.257 | -0.3145 | Yes | ||
| 75 | NDUFS8 | NA | NDUFS8 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) | 24740 | -1.324 | -0.3089 | Yes |
| 76 | ADSL | NA | ADSL Entrez, Source | adenylosuccinate lyase | 25220 | -1.509 | -0.3118 | Yes |
| 77 | MIA3 | NA | MIA3 Entrez, Source | melanoma inhibitory activity family, member 3 | 25545 | -1.661 | -0.3075 | Yes |
| 78 | HSPD1 | NA | HSPD1 Entrez, Source | heat shock 60kDa protein 1 (chaperonin) | 25765 | -1.775 | -0.2982 | Yes |
| 79 | LAP3 | NA | LAP3 Entrez, Source | leucine aminopeptidase 3 | 25812 | -1.803 | -0.2823 | Yes |
| 80 | CSE1L | NA | CSE1L Entrez, Source | CSE1 chromosome segregation 1-like (yeast) | 25848 | -1.822 | -0.2658 | Yes |
| 81 | PDHA1 | NA | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 25871 | -1.839 | -0.2486 | Yes |
| 82 | AFG3L2 | NA | AFG3L2 Entrez, Source | AFG3 ATPase family gene 3-like 2 (yeast) | 25938 | -1.879 | -0.2327 | Yes |
| 83 | ALDH7A1 | NA | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 26299 | -2.130 | -0.2251 | Yes |
| 84 | FAM158A | NA | 26463 | -2.266 | -0.2090 | Yes | ||
| 85 | MTO1 | NA | MTO1 Entrez, Source | mitochondrial translation optimization 1 homolog (S. cerevisiae) | 26598 | -2.380 | -0.1907 | Yes |
| 86 | HLA-B | NA | HLA-B Entrez, Source | major histocompatibility complex, class I, B | 26685 | -2.482 | -0.1696 | Yes |
| 87 | CTSC | NA | CTSC Entrez, Source | cathepsin C | 26690 | -2.488 | -0.1455 | Yes |
| 88 | CORO1B | NA | CORO1B Entrez, Source | coronin, actin binding protein, 1B | 26738 | -2.548 | -0.1223 | Yes |
| 89 | CCT4 | NA | CCT4 Entrez, Source | chaperonin containing TCP1, subunit 4 (delta) | 26763 | -2.581 | -0.0980 | Yes |
| 90 | MRPS30 | NA | MRPS30 Entrez, Source | mitochondrial ribosomal protein S30 | 26847 | -2.746 | -0.0742 | Yes |
| 91 | C12orf11 | NA | C12ORF11 Entrez, Source | chromosome 12 open reading frame 11 | 26996 | -3.035 | -0.0500 | Yes |
| 92 | TNNT1 | NA | TNNT1 Entrez, Source | troponin T type 1 (skeletal, slow) | 27044 | -3.163 | -0.0209 | Yes |
| 93 | PERP | NA | PERP Entrez, Source | PERP, TP53 apoptosis effector | 27060 | -3.208 | 0.0099 | Yes |

