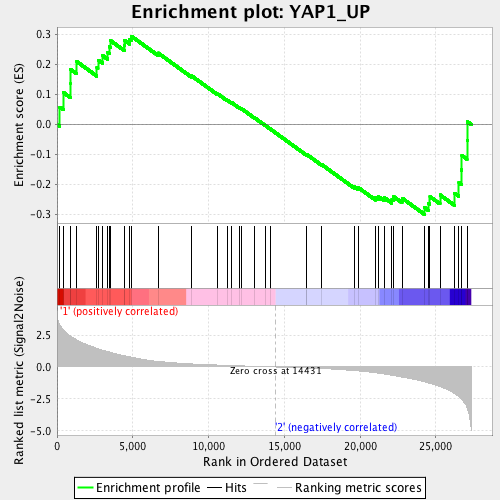
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | YAP1_UP |
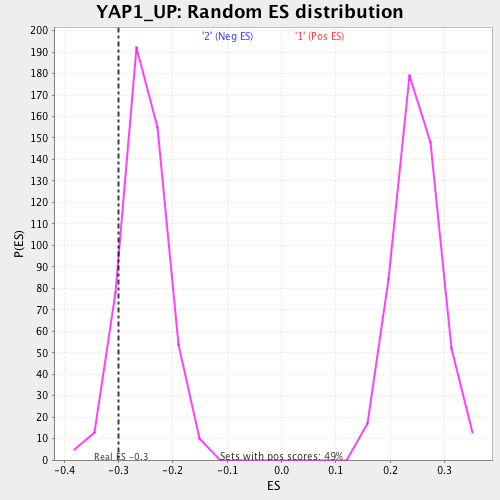
| Enrichment Score (ES) | -0.30026236 |
| Normalized Enrichment Score (NES) | -1.1853057 |
| Nominal p-value | 0.112426035 |
| FDR q-value | 0.23159659 |
| FWER p-Value | 0.925 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SHISA4 | NA | 183 | 3.295 | 0.0581 | Yes | ||
| 2 | C13orf15 | NA | C13ORF15 Entrez, Source | chromosome 13 open reading frame 15 | 413 | 2.909 | 0.1069 | Yes |
| 3 | DDB2 | NA | DDB2 Entrez, Source | damage-specific DNA binding protein 2, 48kDa | 891 | 2.397 | 0.1366 | Yes |
| 4 | CNP | NA | CNP Entrez, Source | 2',3'-cyclic nucleotide 3' phosphodiesterase | 898 | 2.390 | 0.1833 | Yes |
| 5 | RHOQ | NA | RHOQ Entrez, Source | ras homolog gene family, member Q | 1302 | 2.100 | 0.2099 | Yes |
| 6 | MOCS2 | NA | MOCS2 Entrez, Source | molybdenum cofactor synthesis 2 | 2627 | 1.450 | 0.1899 | Yes |
| 7 | MAML2 | NA | MAML2 Entrez, Source | mastermind-like 2 (Drosophila) | 2698 | 1.423 | 0.2153 | Yes |
| 8 | FAM45A | NA | FAM45A Entrez, Source | family with sequence similarity 45, member A | 3009 | 1.307 | 0.2296 | Yes |
| 9 | FLOT1 | NA | FLOT1 Entrez, Source | flotillin 1 | 3347 | 1.200 | 0.2409 | Yes |
| 10 | SIAE | NA | SIAE Entrez, Source | sialic acid acetylesterase | 3474 | 1.162 | 0.2591 | Yes |
| 11 | KIAA1033 | NA | KIAA1033 Entrez, Source | KIAA1033 | 3506 | 1.153 | 0.2806 | Yes |
| 12 | FAF2 | NA | 4435 | 0.875 | 0.2638 | Yes | ||
| 13 | TEAD1 | NA | TEAD1 Entrez, Source | TEA domain family member 1 (SV40 transcriptional enhancer factor) | 4457 | 0.871 | 0.2802 | Yes |
| 14 | C8orf58 | NA | C8ORF58 Entrez, Source | chromosome 8 open reading frame 58 | 4779 | 0.794 | 0.2840 | Yes |
| 15 | RPN2 | NA | RPN2 Entrez, Source | ribophorin II | 4893 | 0.768 | 0.2950 | Yes |
| 16 | PTPMT1 | NA | PTPMT1 Entrez, Source | protein tyrosine phosphatase, mitochondrial 1 | 6662 | 0.433 | 0.2387 | No |
| 17 | CYP4B1 | NA | CYP4B1 Entrez, Source | cytochrome P450, family 4, subfamily B, polypeptide 1 | 8869 | 0.238 | 0.1625 | No |
| 18 | SAMSN1 | NA | SAMSN1 Entrez, Source | SAM domain, SH3 domain and nuclear localisation signals, 1 | 10602 | 0.151 | 0.1020 | No |
| 19 | MSN | NA | MSN Entrez, Source | moesin | 11240 | 0.121 | 0.0811 | No |
| 20 | PRKACG | NA | PRKACG Entrez, Source | protein kinase, cAMP-dependent, catalytic, gamma | 11521 | 0.110 | 0.0730 | No |
| 21 | DLX5 | NA | DLX5 Entrez, Source | distal-less homeobox 5 | 12050 | 0.088 | 0.0553 | No |
| 22 | GTPBP3 | NA | GTPBP3 Entrez, Source | GTP binding protein 3 (mitochondrial) | 12181 | 0.084 | 0.0522 | No |
| 23 | SNX7 | NA | SNX7 Entrez, Source | sorting nexin 7 | 13015 | 0.052 | 0.0227 | No |
| 24 | CCDC8 | NA | CCDC8 Entrez, Source | coiled-coil domain containing 8 | 13765 | 0.024 | -0.0043 | No |
| 25 | KCNK12 | NA | KCNK12 Entrez, Source | potassium channel, subfamily K, member 12 | 14056 | 0.013 | -0.0146 | No |
| 26 | GLTSCR1 | NA | GLTSCR1 Entrez, Source | glioma tumor suppressor candidate region gene 1 | 16477 | -0.080 | -0.1018 | No |
| 27 | RAB40C | NA | RAB40C Entrez, Source | RAB40C, member RAS oncogene family | 16478 | -0.080 | -0.1002 | No |
| 28 | LLGL1 | NA | LLGL1 Entrez, Source | lethal giant larvae homolog 1 (Drosophila) | 17442 | -0.122 | -0.1331 | No |
| 29 | DRAM2 | NA | 19610 | -0.282 | -0.2070 | No | ||
| 30 | SNAP23 | NA | SNAP23 Entrez, Source | synaptosomal-associated protein, 23kDa | 19869 | -0.307 | -0.2104 | No |
| 31 | CXorf56 | NA | CXORF56 Entrez, Source | chromosome X open reading frame 56 | 21008 | -0.453 | -0.2432 | No |
| 32 | ARHGEF10 | NA | ARHGEF10 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 10 | 21221 | -0.484 | -0.2415 | No |
| 33 | COX8A | NA | COX8A Entrez, Source | cytochrome c oxidase subunit 8A (ubiquitous) | 21581 | -0.549 | -0.2438 | No |
| 34 | FLJ27352 | NA | FLJ27352 Entrez, Source | - | 22093 | -0.646 | -0.2498 | No |
| 35 | SEMA3C | NA | SEMA3C Entrez, Source | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | 22187 | -0.662 | -0.2402 | No |
| 36 | FAM35A | NA | FAM35A Entrez, Source | family with sequence similarity 35, member A | 22760 | -0.786 | -0.2457 | No |
| 37 | FAM167A | NA | 24249 | -1.165 | -0.2774 | No | ||
| 38 | YWHAZ | NA | YWHAZ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 24505 | -1.244 | -0.2622 | No |
| 39 | C8orf48 | NA | C8ORF48 Entrez, Source | chromosome 8 open reading frame 48 | 24603 | -1.277 | -0.2407 | No |
| 40 | NRAS | NA | NRAS Entrez, Source | neuroblastoma RAS viral (v-ras) oncogene homolog | 25268 | -1.528 | -0.2350 | No |
| 41 | MYLK | NA | MYLK Entrez, Source | myosin, light chain kinase | 26239 | -2.081 | -0.2296 | No |
| 42 | CDIPT | NA | CDIPT Entrez, Source | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) | 26514 | -2.308 | -0.1942 | No |
| 43 | C9orf95 | NA | C9ORF95 Entrez, Source | chromosome 9 open reading frame 95 | 26672 | -2.465 | -0.1515 | No |
| 44 | ZNHIT6 | NA | 26711 | -2.521 | -0.1033 | No | ||
| 45 | RFTN1 | NA | RFTN1 Entrez, Source | raftlin, lipid raft linker 1 | 27049 | -3.181 | -0.0531 | No |
| 46 | CAMK4 | NA | CAMK4 Entrez, Source | calcium/calmodulin-dependent protein kinase IV | 27064 | -3.222 | 0.0097 | No |